8 辅助工具使用教程
备注
用DSPAW完成DFT计算后,想快速分析结果、画图,或者完成一些常见的数据处理任务?
dspawpy ( Python ≥ 3.8 )就是这样一款辅助工具,它可以编程调用(参考下文示例脚本),也提供了一个命令行交互程序
参考下方教程 安装 后,命令行输入 dspawpy 后回车即可使用交互程序:
... loading dspawpy cli ...
********这是dspawpy命令行交互小工具,预祝您使用愉快********
( )
_| | ___ _ _ _ _ _ _ _ _ _ _ _
/'_` |/',__)( '_`\ /'_` )( ) ( ) ( )( '_`\ ( ) ( )
( (_| |\__, \| (_) )( (_| || \_/ \_/ || (_) )| (_) |
\__,_)(____/| ,__/'`\__,_) \___x___/ | ,__/ \__, |
| | | | ( )_| |
(_) (_) \___/
[版本号]: [安装路径]
======================================
| 1: update更新
| 2: structure结构转化
| 3: volumetricData数据处理
| 4: band能带数据处理
| 5: dos态密度数据处理
| 6: bandDos能带和态密度共同显示
| 7: optical光学性质数据处理
| 8: neb过渡态计算数据处理
| 9: phonon声子计算数据处理
| 10: aimd分子动力学模拟数据处理
| 11: Polarization铁电极化数据处理
| 12: ZPE零点振动能数据处理
| 13: TS的热校正能
|
| q: 退出
======================================
--> 输入数字后回车选择功能:
亮点:
自动补全: 按Tab键即可生效,有助于快速且正确地输入程序所需要的参数
多线程懒加载:在等待用户输入时后台加载模块,大幅缩短等待时间;仅加载必要模块,减少内存占用
注意:
在远程服务器上使用时,由于机器的磁盘读写性能不佳,可能需要较长的启动时间,极端情况下可能长达半分钟(与服务器当时的磁盘读写性能直接相关)。如果无法忍受,请在自己的电脑上安装dspawpy使用
输入 dspawpy 回车后,python会先加载内建模块,完成后显示 … loading dspawpy cli … 提示语句,表明进入第二阶段(开始加载第三方依赖库)
等待第二阶段完成后,会显示欢迎界面,此时dspwapy已完成前期加载,进入第三阶段。后续将动态根据选择的功能模块加载相应的依赖库,从而最大限度减少等待时间
8.1 安装与更新
在HZW机器上,已提前安装 dspawpy,使用以下命令激活虚拟环境即可使用:
source /data/hzwtech/profile/dspawpy.env
在其他机器上,请自行安装 dspawpy(下列两种方式二选一):
使用 mamba 或 conda ,安装包可前往 https://conda-forge.org/download/ 下载
mamba install dspawpy -c conda-forge
#conda install dspawpy -c conda-forge
或使用 pip3 (部分操作系统中没有pip3可执行程序,此时请尝试pip)
pip3 install dspawpy
关于如何设置pip和conda镜像地址以加速安装过程,可参考 https://mirrors.tuna.tsinghua.edu.cn/help/pypi/ 和 https://mirrors.tuna.tsinghua.edu.cn/help/anaconda/
如果安装依然失败,请尝试上面的 mamba/conda 安装法。
警告
在集群上,由于权限问题,公共路径中的pip很可能不支持全局安装python库,必须在 pip install 后面追加 --user 选项安装到家目录 ~/.local/lib/python3.x/site-packages/ 下,其中3.x表示python解释器版本,x可能是8-12之间的任意整数
python将优先读取家目录下的dspawpy,即使公共环境中的dspawpy的版本比家目录中的dspawpy的版本新!
因此,如果以前用 --user 安装过dspawpy,又忘记手动更新家目录下的老版本dspawpy,即使source了公共环境,也无法调用公共环境中的dspawpy,依旧会使用老版本dspawpy,导致一些BUG。
因此,鉴于HZW集群每周自动更新dspawpy,建议不要在家目录下重复安装,已安装的建议删除 。如果在其他集群上,请确保及时手动更新家目录下的 dspawpy
如果不想删除家目录中的dspawpy,又不想更新它,可以在运行python脚本时,尝试 -s 选项避免导入家目录中的 dspawpy: python -s your-script.py。
更新 dspawpy
如果 dspawpy 是通过 mamba/conda 安装的,使用以下命令更新:
mamba update dspawpy
#conda update dspawpy
如果 dspawpy 是通过 pip 安装的,那么:
pip install dspawpy -U # -U 表示安装最新版
备注
如果 pip 使用了国内的镜像网站,可能由于镜像网站尚未同步最新版 dspawpy 而导致无法顺利升级,请使用如下命令告诉 pip 从pypi官网下载安装:
pip install dspawpy -i https://pypi.org/simple --user -U # -i 指定下载地址,--user 表示仅为当前用户安装,-U 表示安装最新版
如果运行时出现 dspawpy 相关 错误信息,请先检查是否已正确导入最新版本的 dspawpy 并检查安装路径:
$ python3 # 或 python
>>> import dspawpy
>>> dspawpy.__version__ # 将输出版本号
>>> dspawpy.__file__ # 将输出安装路径
8.2 structure结构转化
读取结构信息使用 read 函数;将结构信息写入文件,使用 write 函数;快速结构转化,使用 convert 函数:
可参考 2conversion.py 脚本完成转化:
convert 函数的几个关键参数设置规则见下表:
infmt(输入文件格式) |
infile(输入文件名模糊匹配) |
outfmt(输出文件格式) |
outfile(输出文件名模糊匹配) |
说明 |
|---|---|---|---|---|
h5 |
*.h5 |
X |
X |
DS-PAW计算完成后保存的hdf5类型文件 |
json |
*.json |
json |
*.json |
DS-PAW计算完成后保存的json类型文件 |
pdb |
*.pdb |
pdb |
*.pdb |
Protein Data Bank |
as |
*.as |
as |
*.as |
DS-PAW记载原子坐标等信息的结构文件 |
hzw |
*.hzw |
hzw |
*.hzw |
DeviceStudio默认的结构文件 |
xyz |
*.xyz |
xyz |
*.xyz |
读取时仅支持分子结构的单构型,写入时则是包含晶胞的extended-xyz类型轨迹文件 |
X |
X |
dump |
*.dump |
lammps-dump类型轨迹文件 |
X |
*.cif*/*.mcif* |
cif/mcif |
*.cif*/*.mcif* |
Crystallographic Information File |
X |
*POSCAR*/*CONTCAR*/*.vasp/CHGCAR*/LOCPOT*/vasprun*.xml* |
poscar |
*POSCAR* |
VASP文件 |
X |
*.cssr* |
cssr |
*.cssr* |
Crystal Structure Standard Representation |
X |
*.yaml/*.yml |
yaml/yml |
*.yaml/*.yml |
YAML Ain’t Markup Language |
X |
*.xsf* |
xsf |
*.xsf* |
eXtended Structural Format |
X |
*rndstr.in*/*lat.in*/*bestsqs* |
mcsqs |
*rndstr.in*/*lat.in*/*bestsqs* |
Monte Carlo Special Quasirandom Structure |
X |
inp*.xml/*.in*/inp_* |
fleur-inpgen |
*.in* |
FLEUR 结构文件,须额外安装 pymatgen-io-fleur 库 |
X |
*.res |
res |
*.res |
ShelX res 结构文件 |
X |
*.config*/*.pwmat* |
pwmat |
*.config/*.pwmat |
PWmat 文件 |
X |
X |
prismatic |
*prismatic* |
用于STEM模拟的一种文件格式 |
X |
CTRL* |
X |
X |
Stuttgart LMTO-ASA 文件 |
备注
上述表格中
*号表示任意字符,X表示不支持h5, json, pdb, xyz, dump, CONTCAR等格式支持多个结构组成的轨迹信息(常见于结构优化或者NEB或者AIMD任务)
in(out)fmt 参数优先级高于文件名模糊匹配规则;例如,指定 in(out)fmt=’h5’ 后,文件名可以是任意值,甚至可以是 a.json
将结构信息写为 json 格式时,仅支持可视化 NEB 链任务,详见 观察NEB链 节相关说明
DS-PAW 输出的 neb.h5、phonon.h5、phonon.json、neb.json以及wannier.json 暂时无法读取结构信息
8.3 volumetricData数据处理
volumetricData可视化
差分volumetricData可视化
参考
3dvol.py:上述脚本支持处理多元体系的电荷密度差,示例以二元体系为例,得到了 AB.h5 与 A.h5 和 B.h5 之间的电荷密度差文件 delta_rho.cube ,该文件可直接使用 VESTA 打开。
volumetricData面平均
参考
3planar_ave.py:
处理 应用案例 3.3 小节所得静电势文件,可得真空方向势函数曲线如下所示:
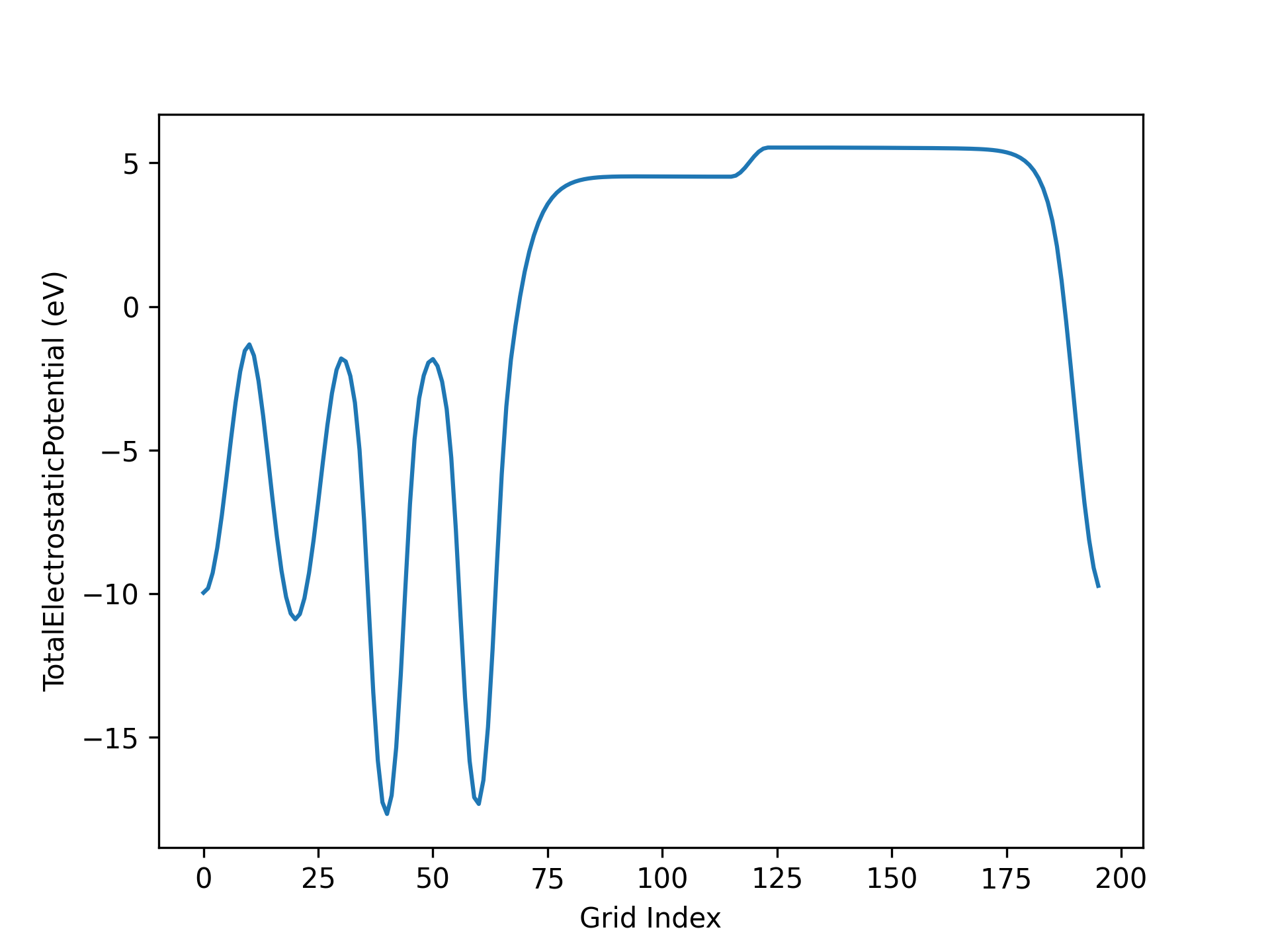
AuAl势函数示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.4 band能带数据处理
知识点:
脚本中调用get_band_data()读取数据,在读取数据时设置efermi=XX可将能量零点修改为指定值;设置zero_to_efermi=True, 可将能量零点修改问所读取的文件中的费米能级处
脚本调用pymatgen的BSPlotter.get_plot()画图,在画图时可设置zero_to_efermi=True,将坐标轴能量零点修改到费米能级处。由于pymatgen在2023.8.17的一处关键更新将绘图函数返回对象从plt改成了axes,导致后续脚本可能出现不兼容。因此用户脚本相关部分已添加一个判断语句加以处理。
能带两步算需从第一步自洽计算获取准确费米能级(从自洽获得的system.json),若获取失败,用户可在调用get_band_data读取数据时,利用参数efermi修改能量零点。例如:band_data=get_band_data(‘band.h5’,efermi=-1.5)
脚本中画图调用pymatgen中的BSPlotter.get_plot, 当判断体系为非金属时,设置zero_to_efermi会认为VBM为费米能级能量而非文件读取时的费米能级。故当体系为非金属时,在数据读取时设置zero_to_efermi=True和在画图时设置zero_to_efermi=True得到的图会有区别
执行本节所列的python脚本,程序会判断是否为金属体系。若为非金属体系,将要求选择是否平移费米能级到能量零点,请按提示操作。
普通能带处理
参考 4bandplot.py :
知识点:
能带两步算需从自洽计算获取准确的费米能级(从system.json获取),若获取失败,用户可在 get_band_data 函数中指定 efermi 参数修改
执行代码可以得到类似以下能带图:
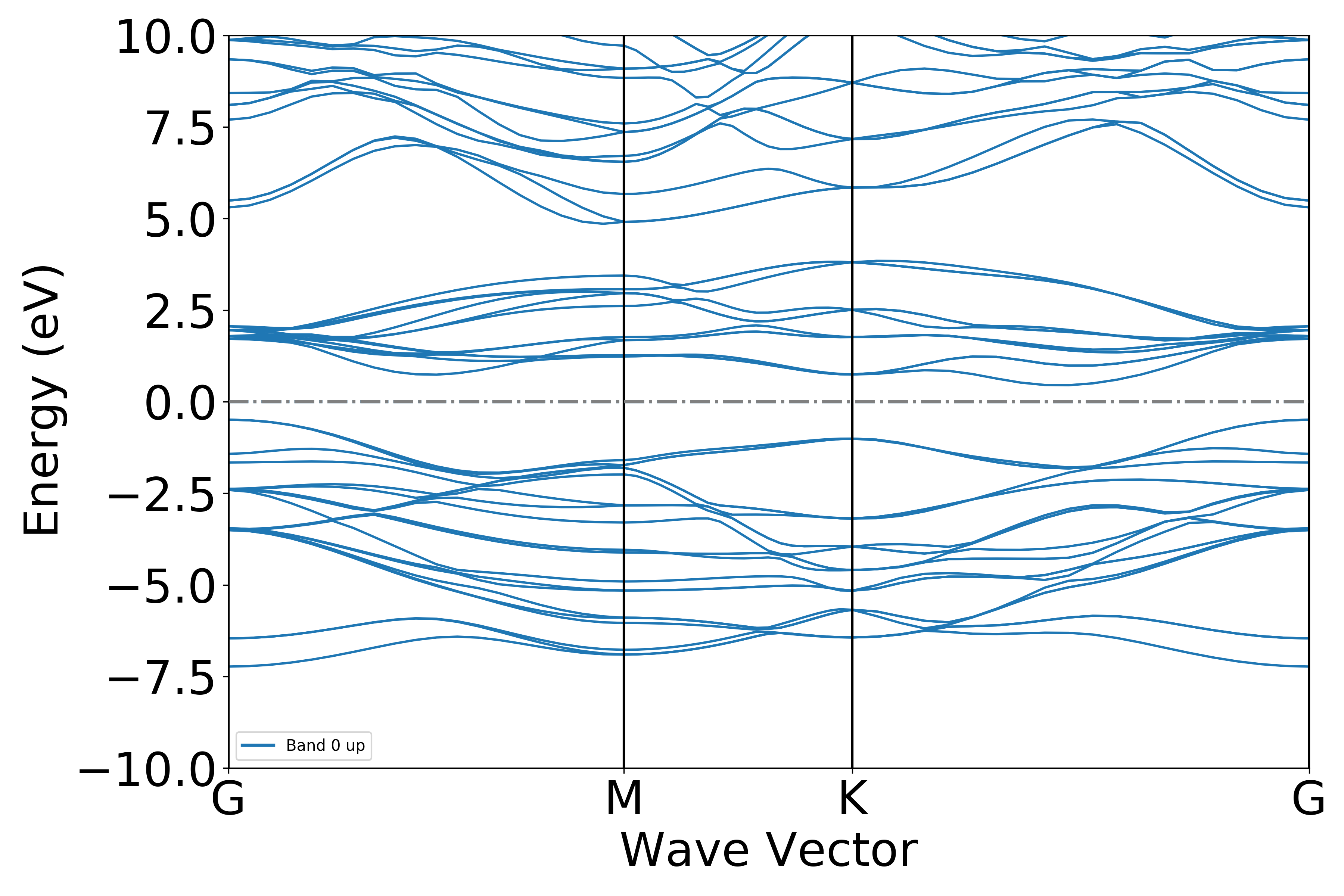
二硫化钼能带示意图
将能带投影到每一种元素分别作图,数据点大小表示该元素对该轨道的贡献
参考 4bandplot_elt.py :
知识点:
用户如果需要绘制能带投影的数据,此时需要使用 BSPlotterProjected模块
使用 BSPlotterProjected模块中 get_elt_projected_plots 函数能够绘制每种元素对轨道贡献的能带图
执行代码可以得到类似以下能带图:
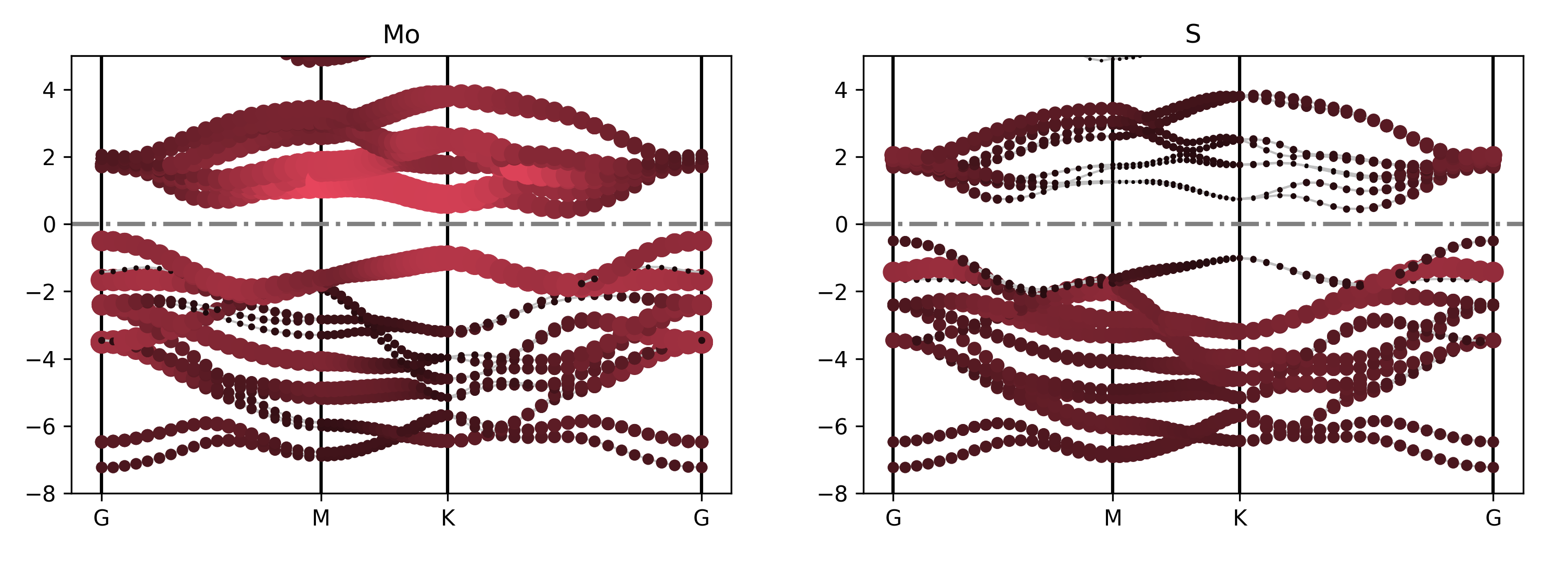
二硫化钼元素投影能带示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
能带投影到不同元素的不同轨道
参考 4bandplot_elt_orbit.py :
知识点:
使用 BSPlotterProjected模块中 get_projected_plots_dots可以让用户来自定义需要绘制的某种元素某种轨道(L)的能带图
例如 get_projected_plots_dots ({‘Mo’:[‘d’],’S’:[‘s’]})就是绘制Mo的d轨道和S的s轨道
执行代码可以得到类似以下能带图:
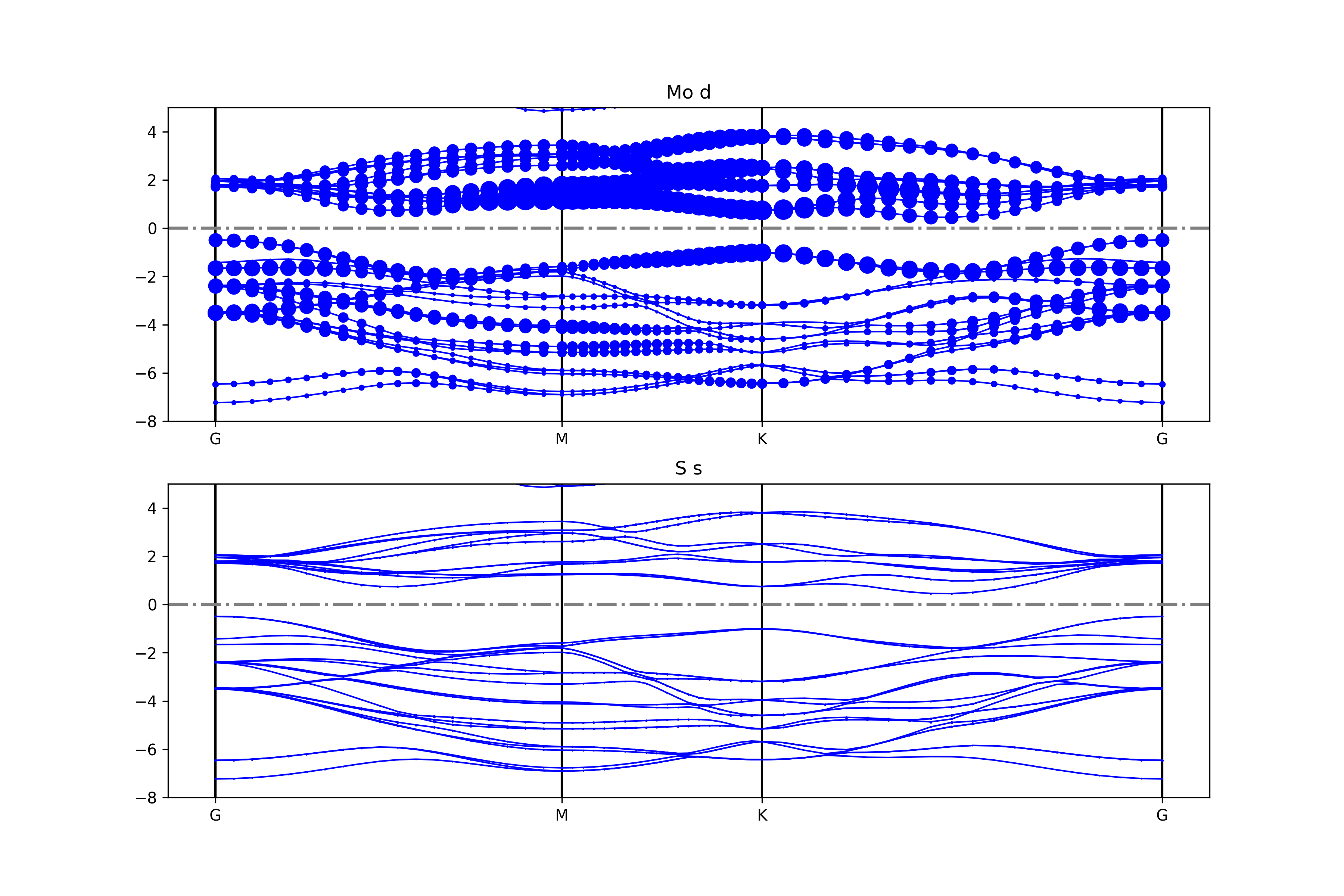
二硫化钼元素轨道投影能带示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
将能带投影到不同原子的不同轨道
参考 4bandplot_patom_porbit.py :
知识点:
使用 BSPlotterProjected模块中 get_projected_plots_dots_patom_pmorb 的自由度更高,可以让用户来自定义需要绘制的某些原子某些轨道的能带图
dictpa指定原子,dictio 指定该原子的轨道
如果要将一些原子或一些轨道的投影分量叠加起来,请根据 get_projected_plots_dots_patom_pmorb 函数文档指定 sum_atoms 或 sum_morbs 参数
警告
如果只选择单个轨道,且轨道名称不止一个字母(例如px、dxy、dxz),get_projected_plots_dots_patom_pmorb 函数将报错,详情见 此处
执行代码可以得到类似以下能带图:
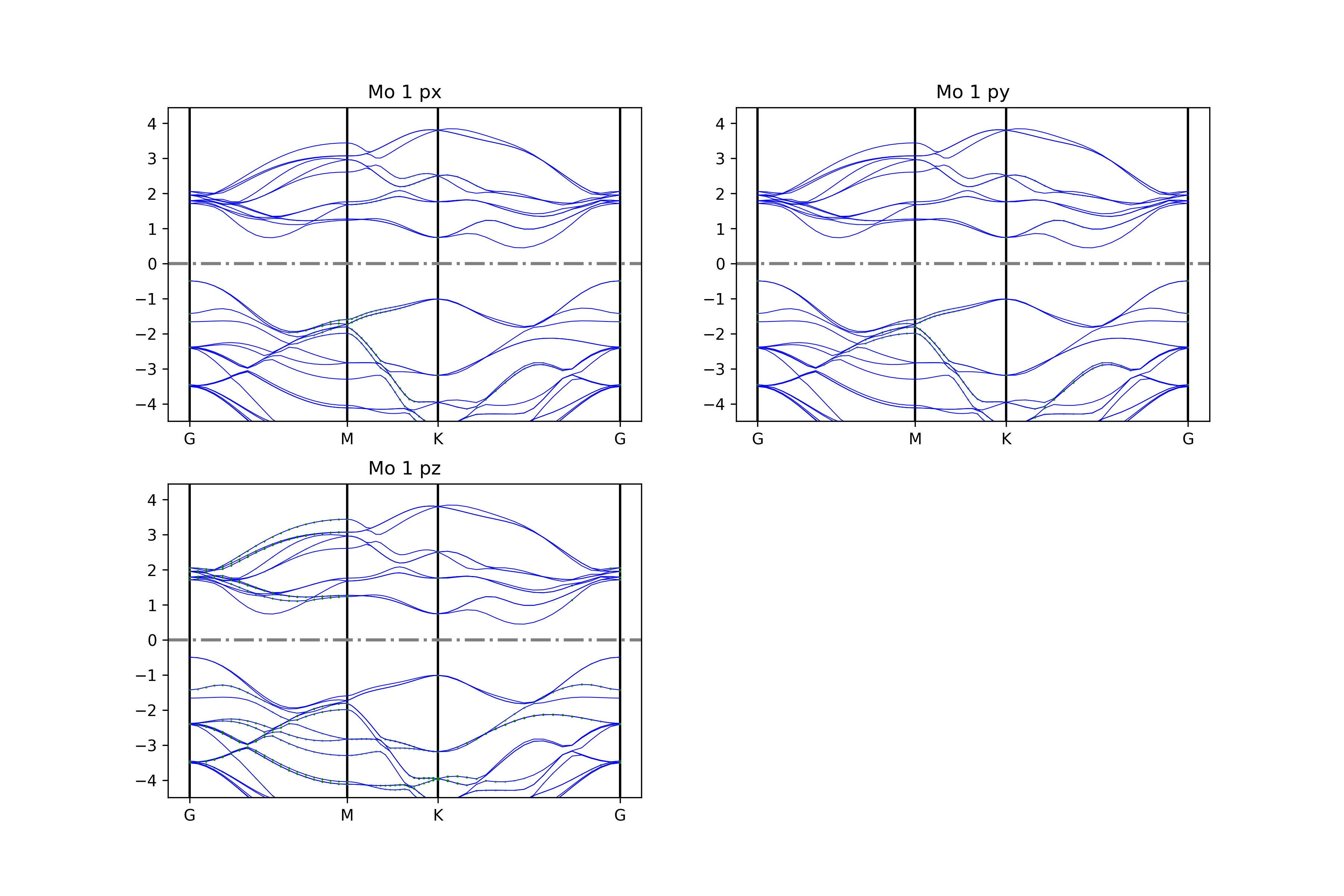
二硫化钼原子投影能带示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
能带反折叠处理
参考 4bandunfolding.py :
执行代码可以得到类似以下能带图:
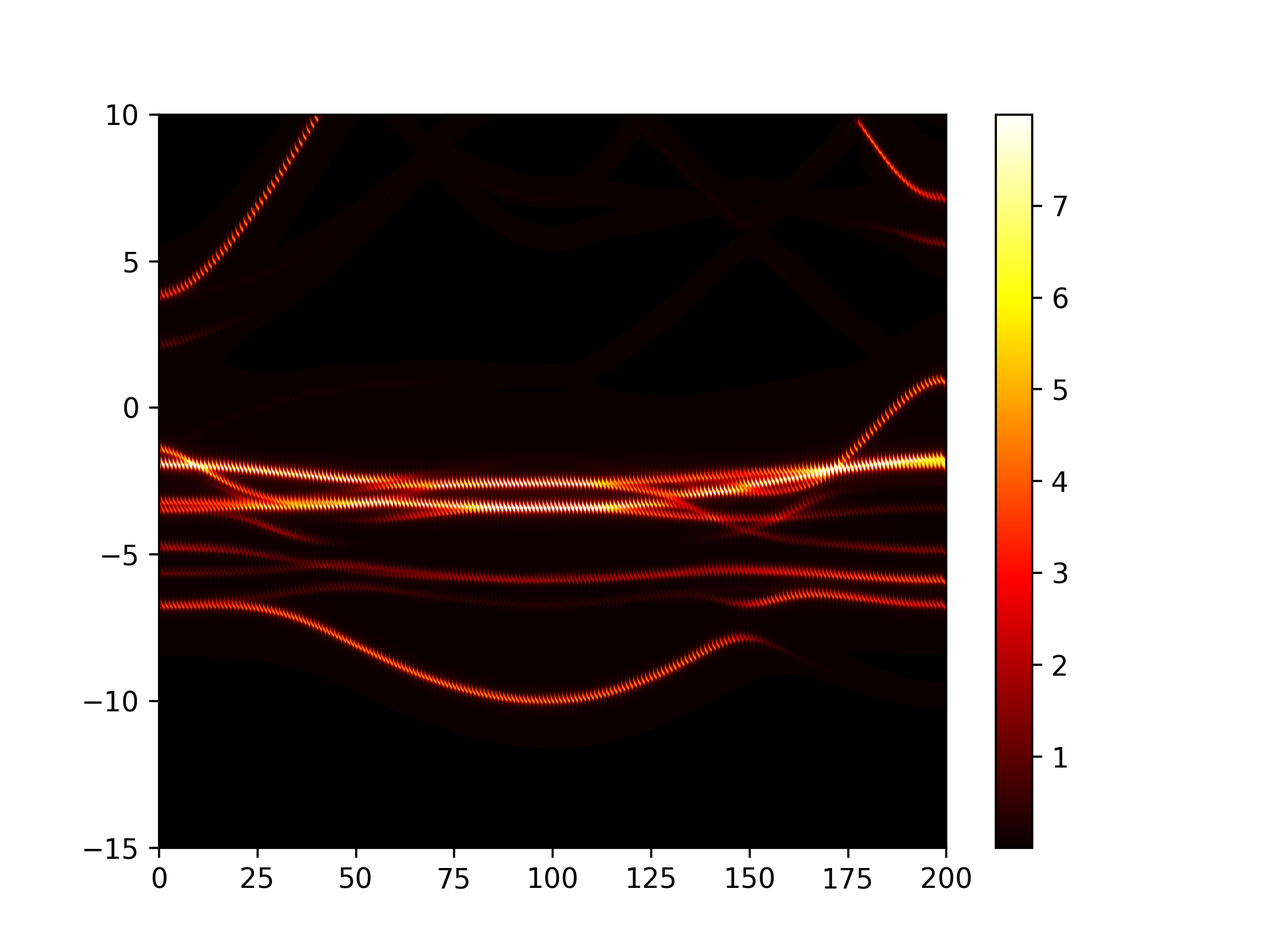
Cu3Au能带反折叠示意图
警告
此功能暂不支持将非金属材料的费米能级设为能量零点(默认价带顶为能量零点)。
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
band-compare能带对比图处理
将普通能带和wannier能带绘制在同一张图上
参考 4bandcompare.py :
执行代码可得到类似如下能带对比曲线:
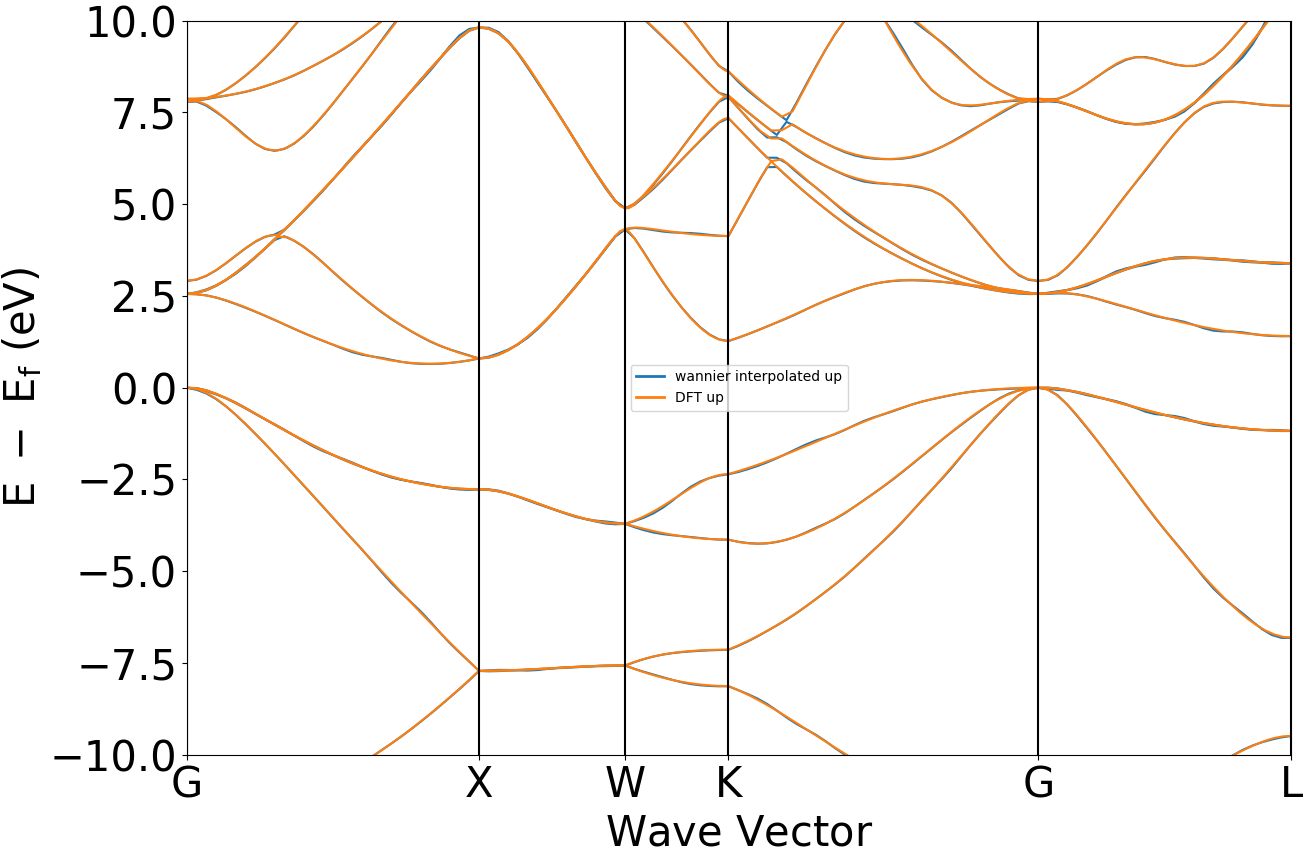
晶体硅瓦尼尔能带与DFT能带示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.5 dos态密度数据处理
总的态密度
参考 5dosplot_total.py :
知识点:
使用 get_dos_data 函数可以将DS-PAW计算得到的 dos.h5 文件转化为 pymatgen 支持的格式
使用 DosPlotter模块获取到DS-PAW计算的 dos.h5 的数据
DosPlotter函数可以传递参数:stack参数表示画态密度是否加阴影, zero_at_efermi 表示是否在态密度图中进行将费米能量置零,这里设置 stack=False , zero_at_efermi=False
使用 DosPlotter 模块中 add_dos 获取态密度的数据
DosPlotter模块中 get_plot函数 绘制态密度图
执行代码可以得到类似以下态密度图:
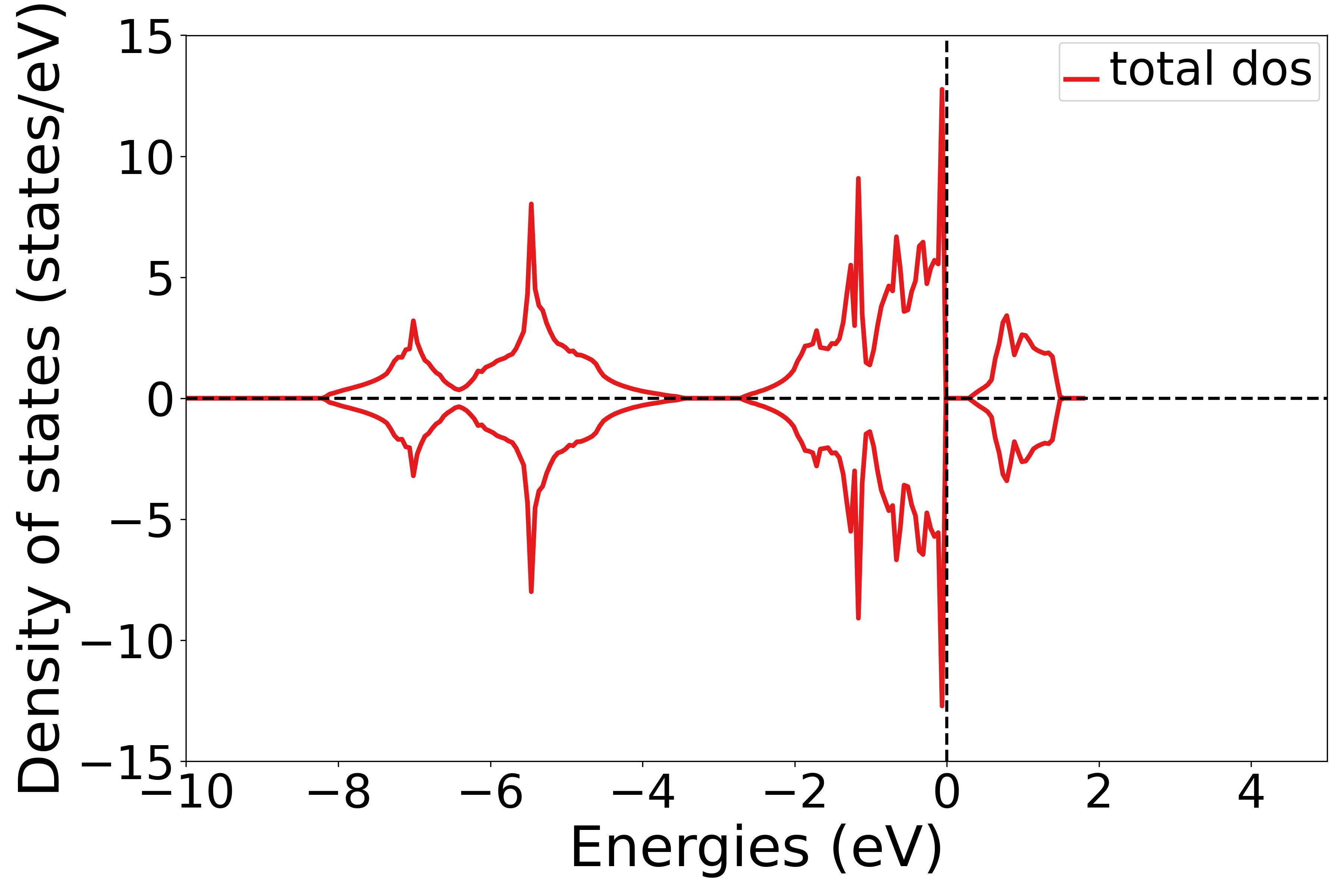
NiO态密度示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
将态密度投影到不同的轨道上
参考 5dosplot_spd.py :
知识点:
使用 DosPlotter模块中 add_dos_dict 函数 获取投影态密度的数据,之后使用 get_spd_dos 将投影信息按照 spd 轨道投影输出
执行代码可以得到类似以下态密度图:
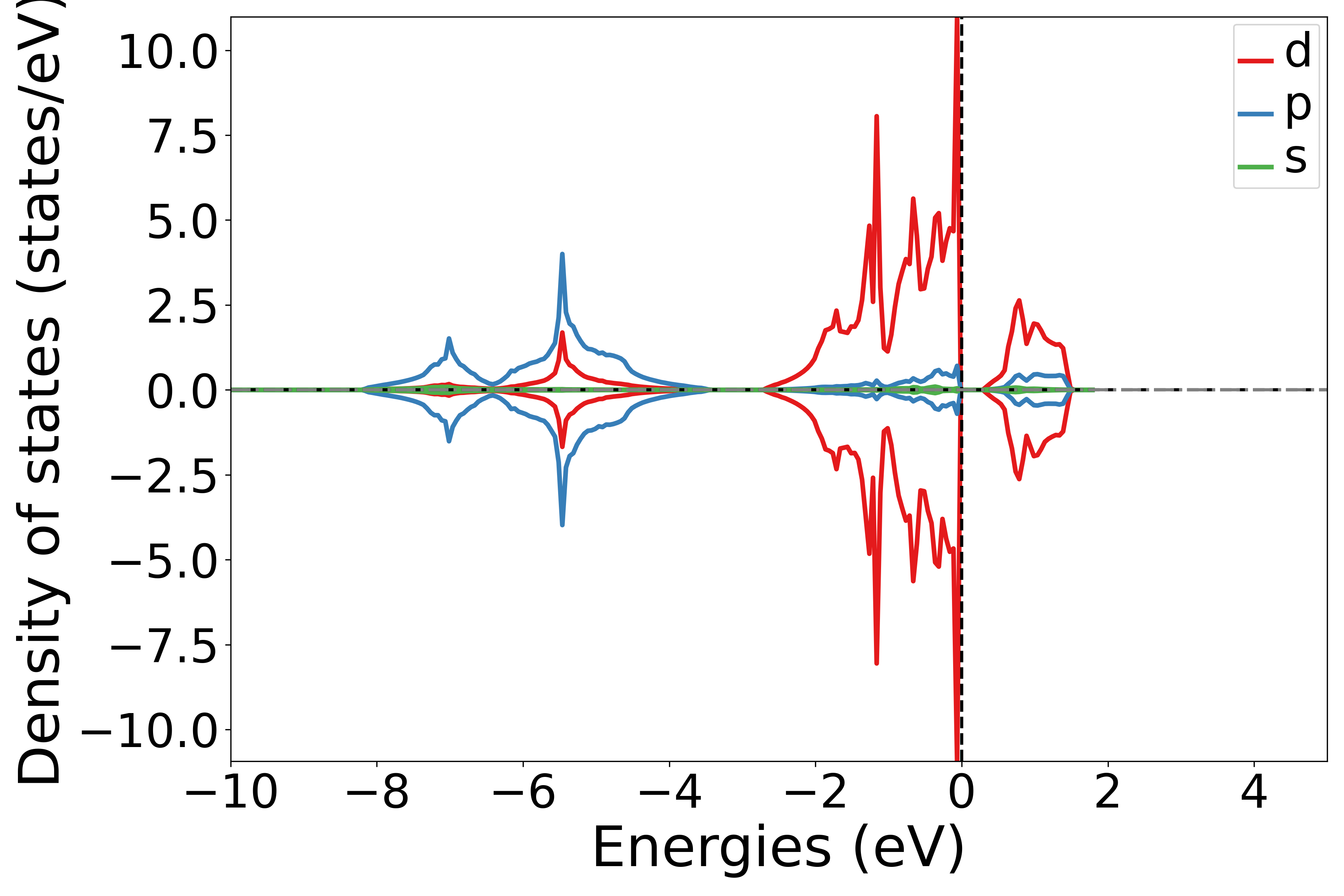
NiO轨道投影态密度示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
将态密度投影到不同的元素上
参考 5dosplot_elt.py :
知识点:
使用 DosPlotter模块中 add_dos_dict 函数 获取投影态密度的数据,之后使用 get_element_dos 将投影信息按照不同元素投影输出
执行代码可以得到类似以下态密度图:
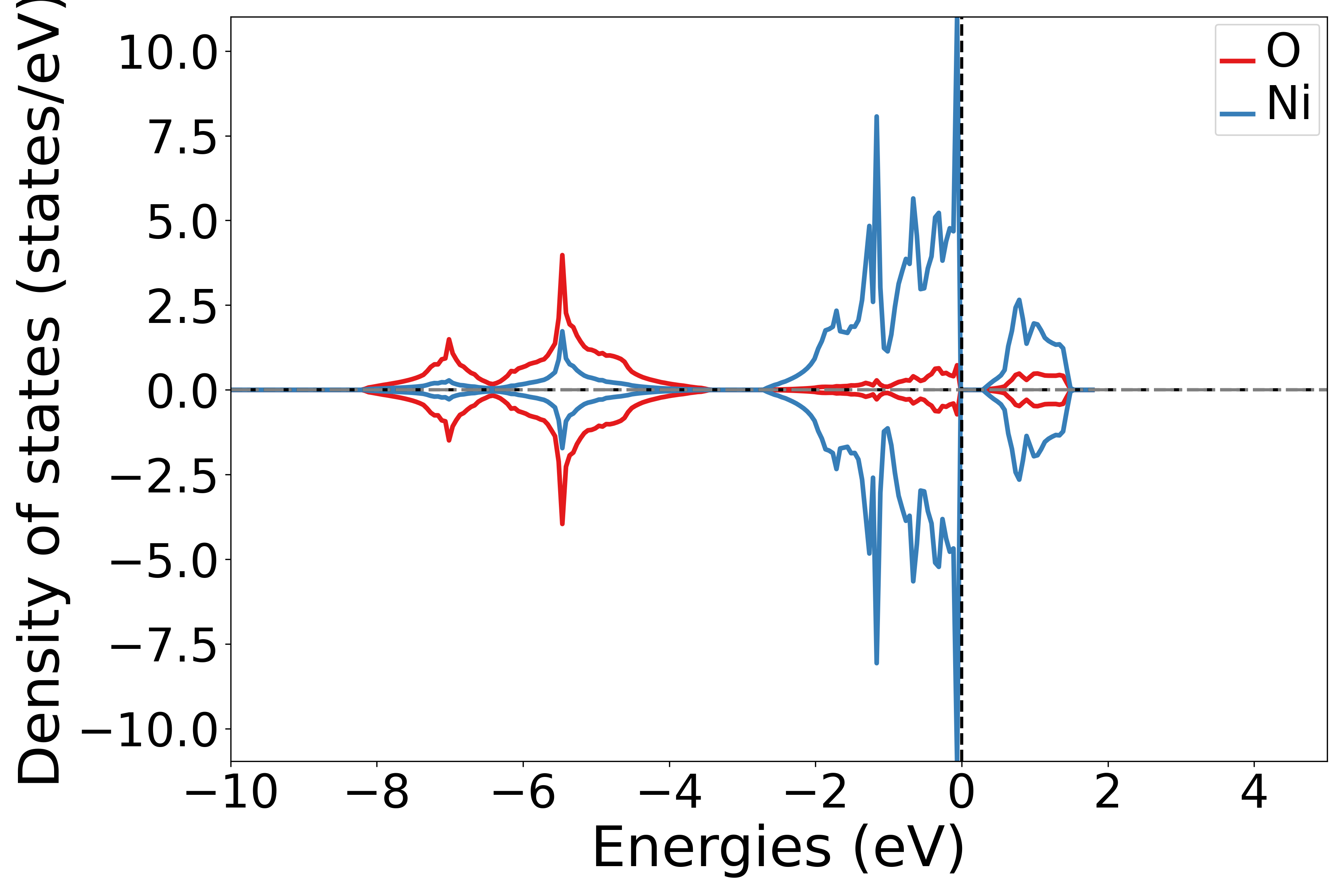
NiO元素投影态密度示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
将态密度投影到不同原子的不同轨道上
参考 5dosplot_atom_orbit.py :
知识点:
使用 get_site_orbital_dos函数提取dos数据中特定原子特定轨道的贡献,dos_data.structure[0],Orbital(4) 表示获取第1个原子的dxy轨道的态密度,get_site_orbital_dos函数中序号从0开始
运行此脚本,根据提示选择元素和轨道,可以得到相应的态密度图
执行代码可以得到类似以下态密度图:
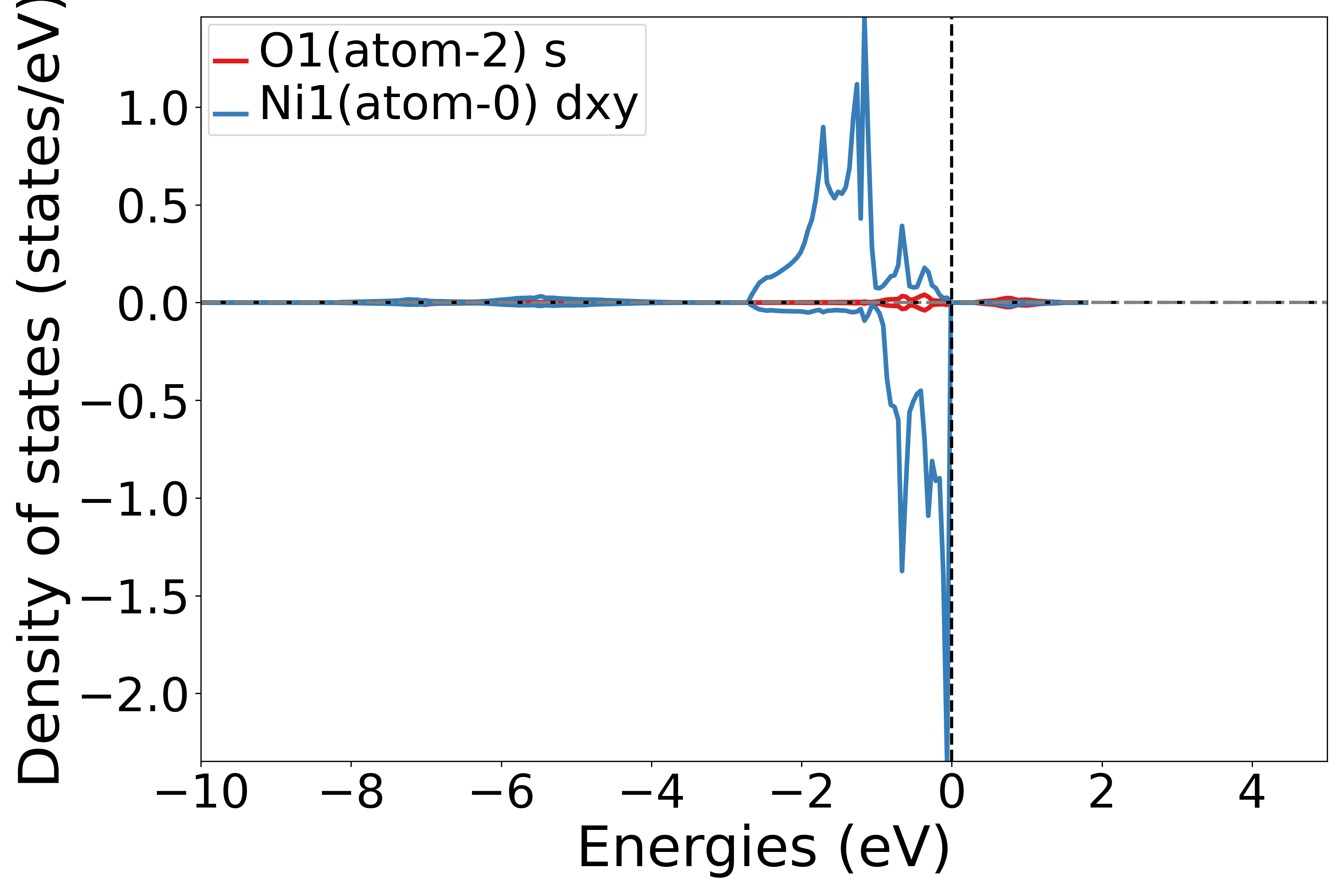
NiO原子轨道态密度示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
将态密度投影到不同原子的分裂d轨道(t2g, eg)上
参考 5dosplot_t2g_eg.py :
知识点:
使用 get_site_t2g_eg_resolved_dos函数 提取dos数据中特定原子的 t2g和 eg轨道的贡献,这是获取第2个原子的t2g和eg轨道的贡献
运行此脚本,根据提示选择原子序号,可以得到相应的态密度图
执行代码可以得到类似以下态密度图:
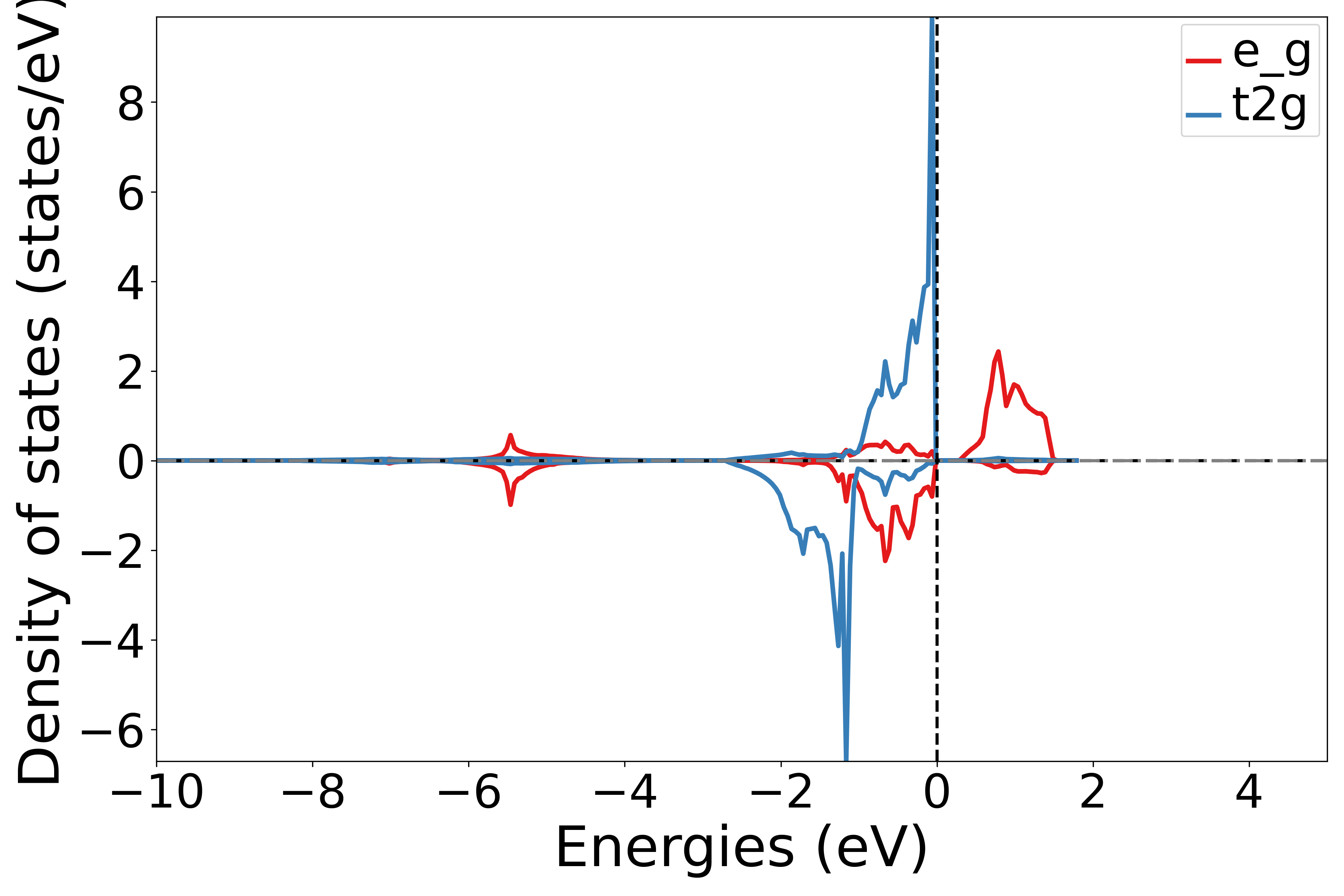
NiO分裂d轨道原子投影态密度示意图
知识点:
如果元素不含d轨道,会画成空白图片
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
d-带中心分析
以 Pb-slab 体系为例,对 Pt 原子进行 d-band 中心分析:
参考 5center_dband.py :
执行代码可以得到类似以下结果:
spin=1
-1.785319344084034
备注
目前仅支持全部原子平均的 d 轨道中心,不支持元素、原子投影或其他轨道,也不支持选择自旋方向或能量范围。
get_dos_data 函数负责处理态密度数据:
8.6 bandDos能带和态密度共同显示
以应用教程中Si体系为例:
将能带和态密度显示在一张图上
参考 6bandDosplot.py :
执行代码可以得到类似以下能带态密度图:
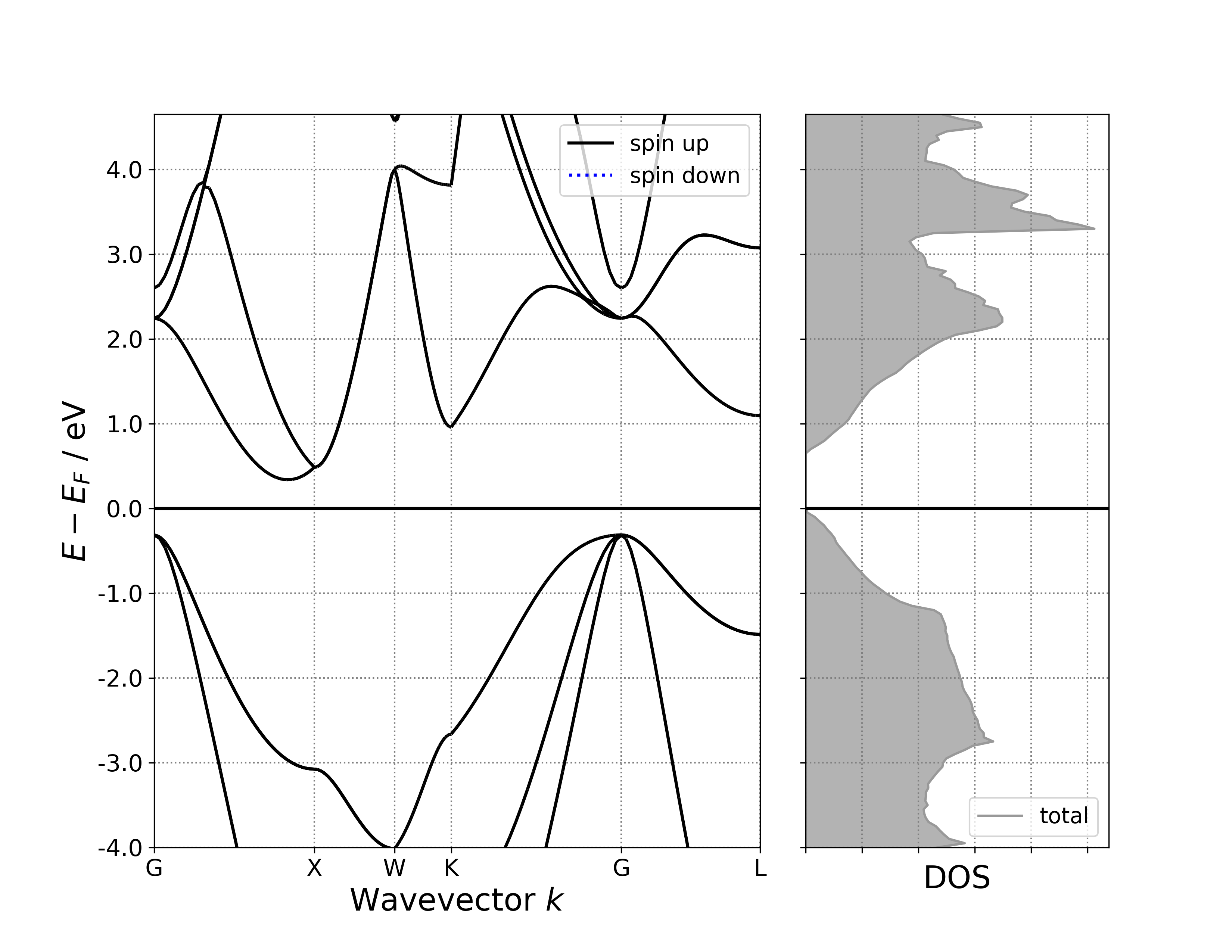
单晶硅能带-态密度示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
将能带和投影态密度显示在一张图上
参考 6bandPdosplot.py :
执行代码可以得到类似以下能带态密度图:
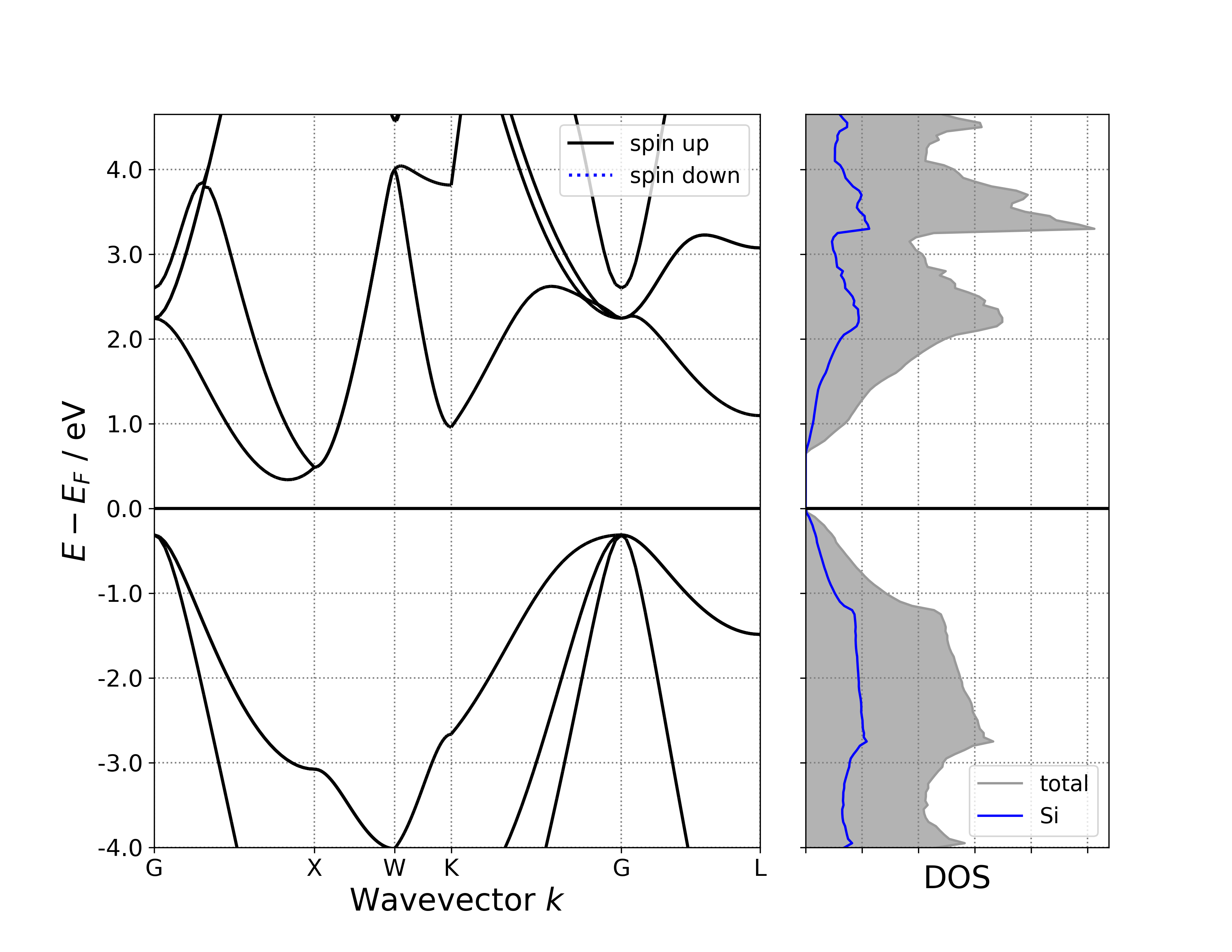
单晶硅能带-投影态密度示意图
警告
给定投影能带数据,默认将沿着元素投影;给定普通能带数据(或体系所含元素种类超过4种),则不投影,并输出警告
给定投影态密度数据,默认也将沿着元素投影,可以换成沿着轨道投影,或者不投影;给定普通态密度数据且未关闭态密度投影选项 BSDOSPlotter(dos_projection=None),pymatgen绘图程序将报错,这就是上面特意准备了一个 6bandDosplot.py 文件的原因。
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.7 optical光学性质数据处理
以快速入门Si体系光学性质计算得到的 scf.h5 为例(注:输出文件名与task一致,task = scf,io.optical = true可计算光学性质):
反射率数据处理,参考 7optical.py :
知识点:
Reflectance为光学性质中的一种,用户可以根据自己的需求将该关键词修改为“AbsorptionCoefficient”或”ExtinctionCoefficient”或”RefractiveIndex”,分别对应吸收系数、消光系数和折射率
执行代码可以得到类似以下反射率随能量变化的曲线:
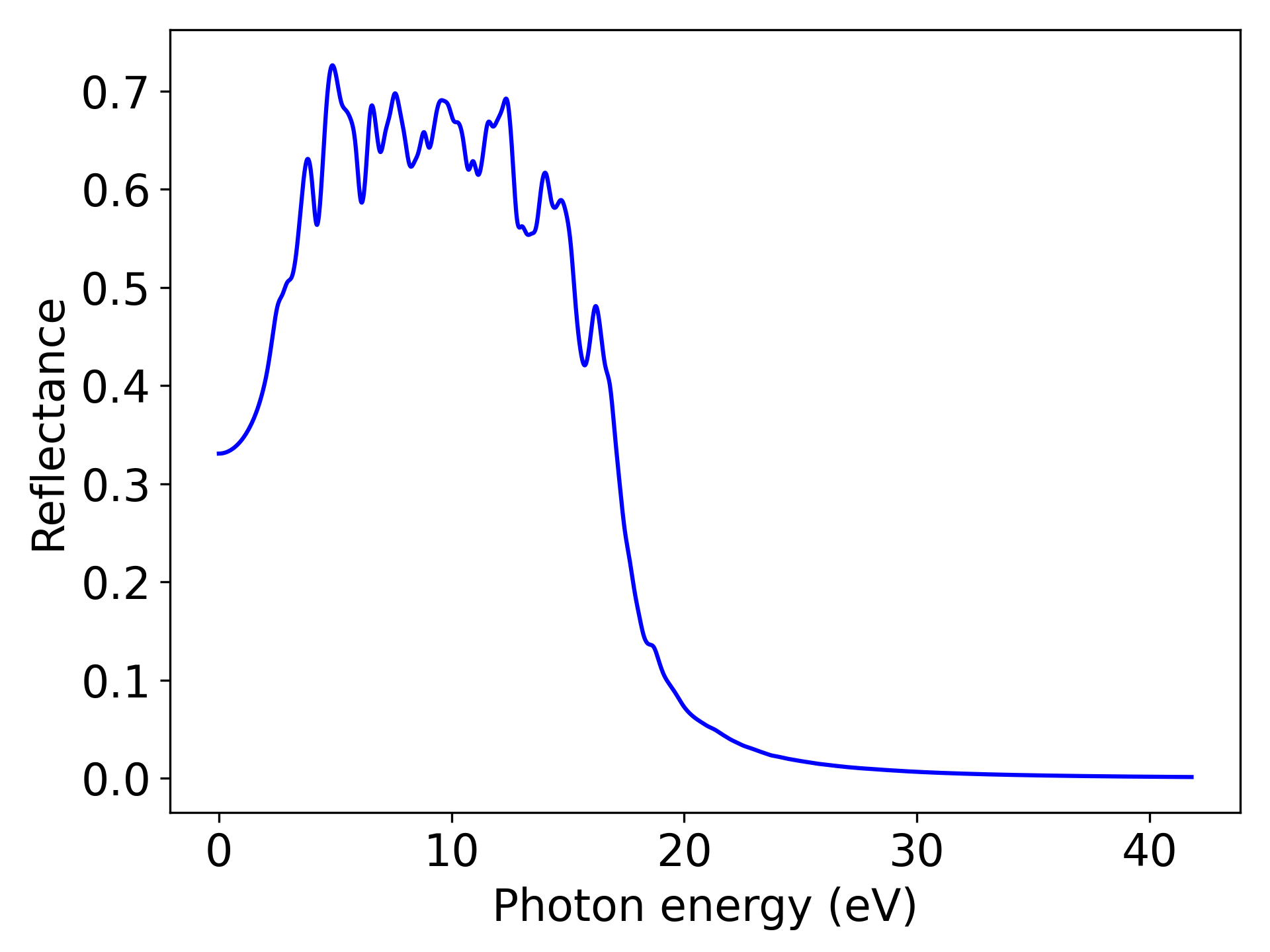
单晶硅Reflectance随光子能量变化趋势示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.8 neb过渡态计算数据处理
以快速入门 H 在 Pt(100) 表面扩散为例进行介绍:
输入文件之生成中间构型
参考
8neb_interpolate_structures.py:
知识点:
用户可以根据需要自行修改插点个数,设置为8得到包含8个结构文件的文件夹,其中中间构型为6个
neb.linear_interpolate为线性插值方法,pbc参数为 True 时将锁定寻找最短扩散路径,默认为 False 以增加用户控制的灵活性,这是因为
举个例子,初态某原子的分数坐标是 0.2,终态变成 0.8。pbc = True 将强制设置扩散路径为 0.2 -> -0.2。pbc = False 则用户可以令程序按照 0.2 -> 0.8 的扩散路径进行插值;如果要用最短路径,手动将 0.8 改为 -0.2 即可,从而确保程序按照使用者的意图完成插值初猜。
绘制能垒图
neb.iniFin = true/false
当设置 neb.iniFin = true/false 时,都可使用读取neb计算的路径进行势垒分析(确保初末态计算文件在neb计算路径下):
参考
8neb_barrier_CubicSpline.py:
备注
运行上述脚本后,可得到类似以下三次样条插值的势垒曲线:
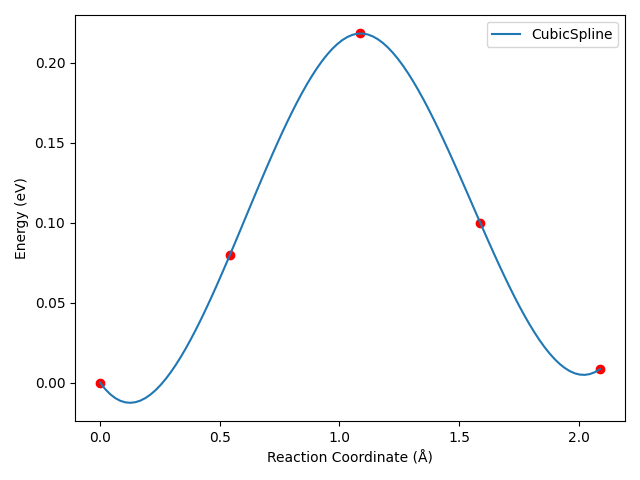
对于这个算例,三次样条插值后,曲线会出现不合理的“下坠”区域,这是三次样条插值算法的特性所决定的。
dspawpy 内部调用 scipy 的插值算法 ,上面这个脚本我们以三次样条插值算法为例,它在 scipy 文档中的定义为:
class scipy.interpolate.CubicSpline(x, y, axis=0, bc_type='not-a-knot', extrapolate=None)
关键词参数包括 axis, bc_type, extrapolate,具体含义见 scipy.interpolate.CubicSpline 。我们可以往 plot_barrier 函数中指定相应的关键词参数(axis, bc_type, extrapolate),将其传递给 scipy.interpolate.CubicSpline 这个类处理。
下面我们采用 8neb_barrier.py 脚本,对比三种算法插值绘制出的曲线:
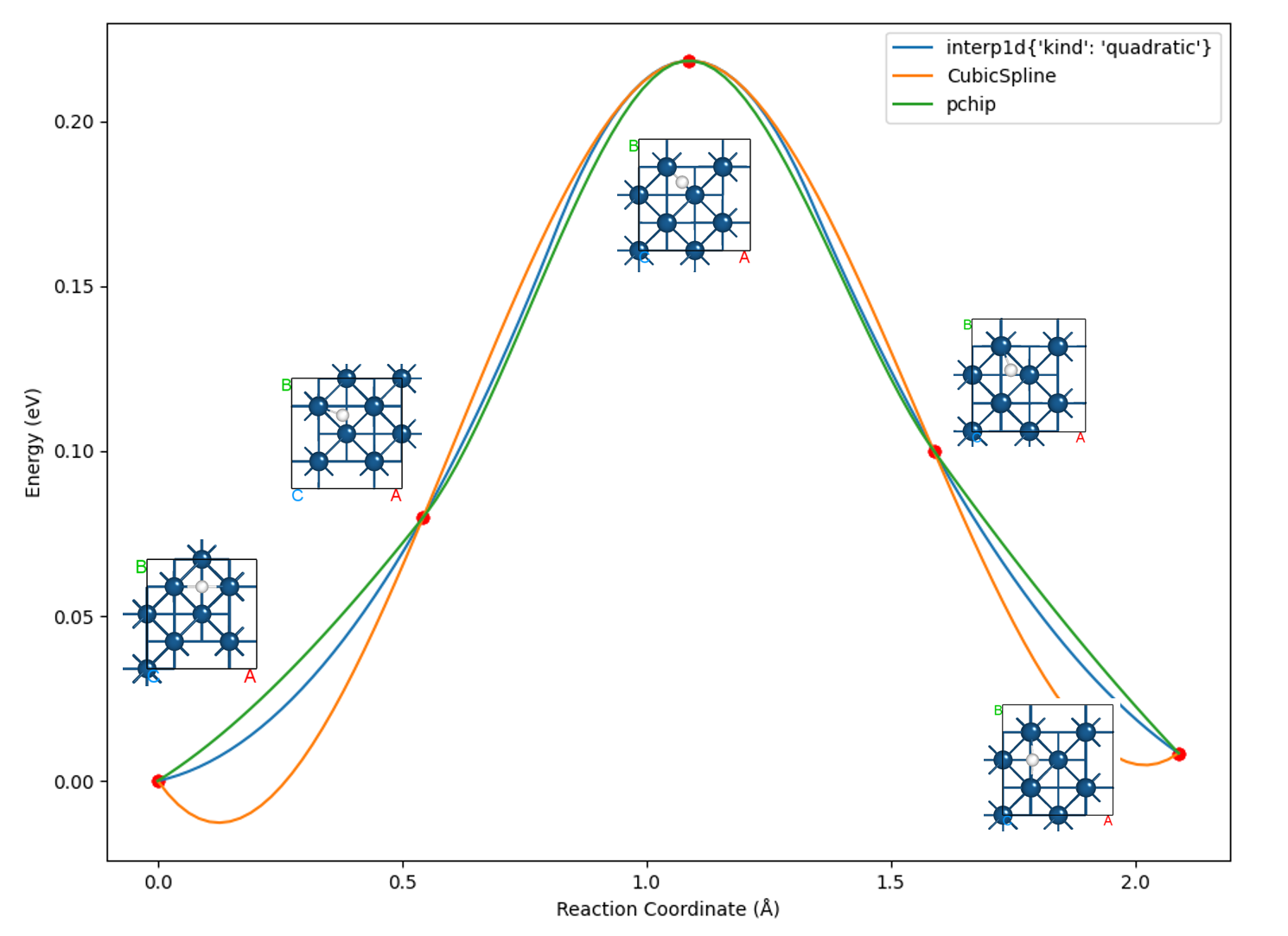
知识点:
选择合适的插值算法对于优化最终曲线的呈现效果至关重要
多数情况下,选择 pchip 这种单调三次样条插值算法即可达到较好效果,这也是默认调用的插值算法
neb.iniFin = true
当设置 neb.iniFin = true 时,读取neb计算所得的 neb.h5/neb.json 文件可快速进行势垒分析:
参考
8neb_barrier_CubicSpline.py:
处理得到的势垒图与之前读取路径效果一致。
备注
neb.h5 和 neb.json文件所存能量为TotalEnergy,如需得精确的势垒值,建议采用读取neb计算路径的方法进行处理(取TotalEnergy0)
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
过渡态计算数据处理
NEB计算完成后,一般要画出能垒图观察,并检查各插值构型最终的受力大小,从而确保受力小于指定的阈值。如果结果异常,还需要检查各插值构型在结构优化过程中的受力与能量变化趋势,判断是否真正“收敛”。上述这些操作至少需要三次循环,为简化操作,我们提供了一个一步到位的总结函数 summary :
参考
8neb_check_results.py:知识点:
此脚本将以表格形式打印各构型的能量和受力、绘制能垒图,并绘制中间构型的能量与受力收敛过程图
若
neb.iniFin = false,用户必须将自洽计算的结果文件 scf.h5 或 system.json 文件复制到对应的初末态子文件夹中,否则程序无法读取初末态能量和受力信息,将报错退出默认情况下,能垒图存储在neb计算目录外层,各中间构型的能量与受力收敛图存储在各子文件夹
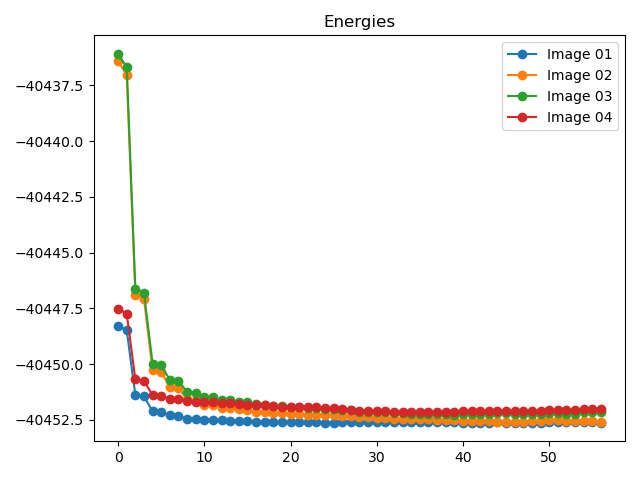
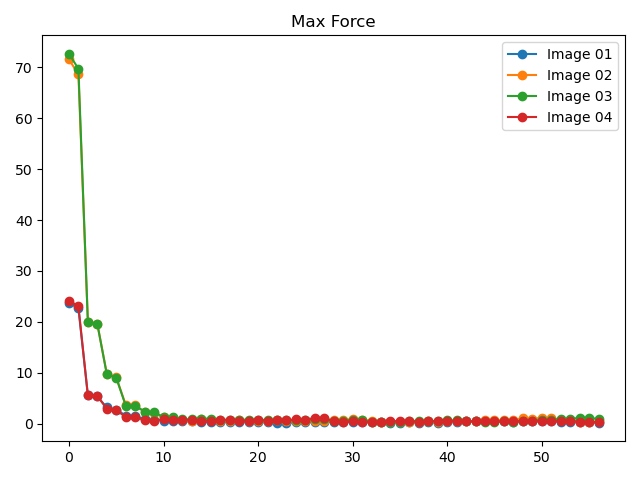
执行代码可以得到类似如下NEB计算各构型的能量和受力表格:
Image
Force (eV/Å)
Reaction coordinate (Å)
Energy (eV)
Delta energy (eV)
00
0.1803
0.0000
-39637.0984
0.0000
01
0.0263
0.5428
-39637.0186
0.0798
02
0.0248
1.0868
-39636.8801
0.2183
03
0.2344
1.5884
-39636.9984
0.1000
04
0.0141
2.0892
-39637.0900
0.0084
除了可以获得能垒图外,还可以得到各中间构型的能量和受力收敛曲线(以02构型为例)
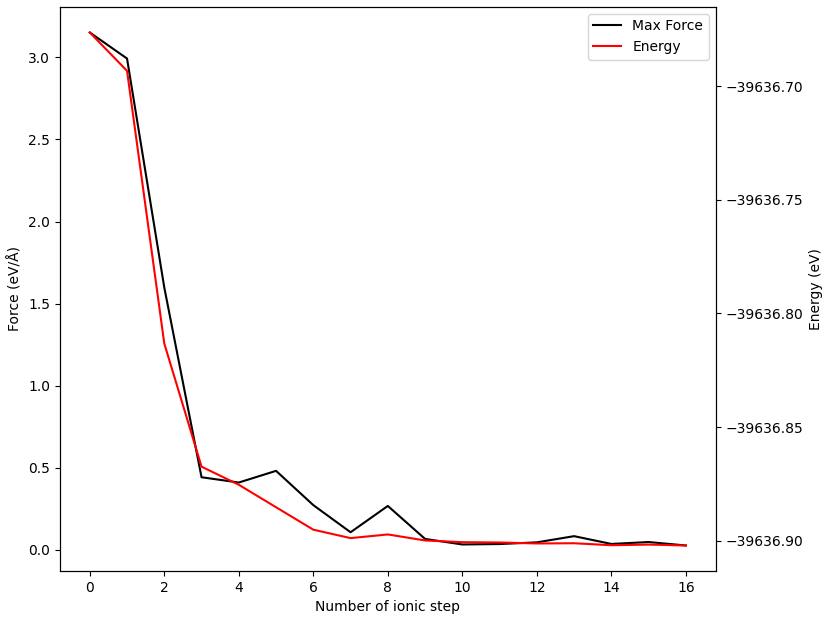
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
观察NEB链
此处所说NEB链指的是各插值构型(structure00.as, structure01.as, …)之间的几何关系,而不是单个构型在优化过程中的变化。
NEB任务计算量较大,观察NEB链有助于判断NEB计算的收敛速度。另外,通过插值算法生成中间结构后,经常需要预览NEB链。这些需求可以通过
8neb_visualize.py脚本实现:
知识点:
此脚本生成neb_movie*.json文件后,通过
Device Studio–>Simulator–>DS-PAW–>Analysis Plot打开 json 文件即可观察directory指定为NEB计算主路径,需提供neb计算完成后的完整文件夹
该脚本支持处理正在进行中的(即未完成的)neb计算文件,方便用户监测实时轨迹
xyz文件可通过 OVITO 软件打开查看:通过
Device Studio–>Simulator–>OVITO打开可视化界面,将xyz文件拖入即可结构信息读取优先级:latestStructureXX.as > h5 > json;设置ignorels为True后,先尝试读取 h5 中的数据,失败则读取 json 中的数据
计算构型间距
参考这个脚本
8calc_dist.py:
neb续算
如果需对neb进行续算,可参考
8neb_restart.py:具体效果见 neb过渡态计算续算说明。
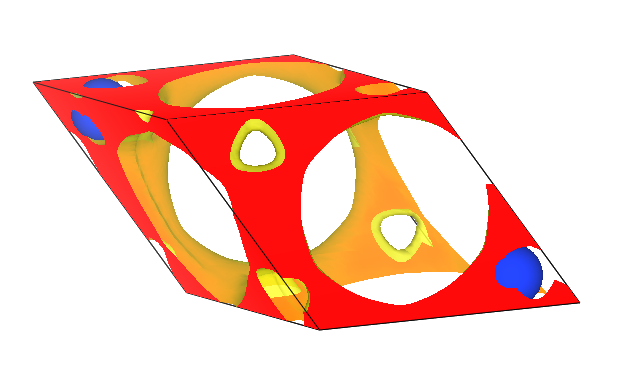
neb计算过程中能量和最大原子受力的变化趋势图
8.9 phonon声子计算数据处理
以MgO体系的声子能带态密度计算得到的 phonon.h5 为例:
如果未安装 phonopy,运行下列脚本时会弹出 no module named 'phonopy' 信息,不影响程序正常运行
声子能带数据处理
参考
9phonon_bandplot.py:
执行代码可以得到类似以下声子能带曲线:
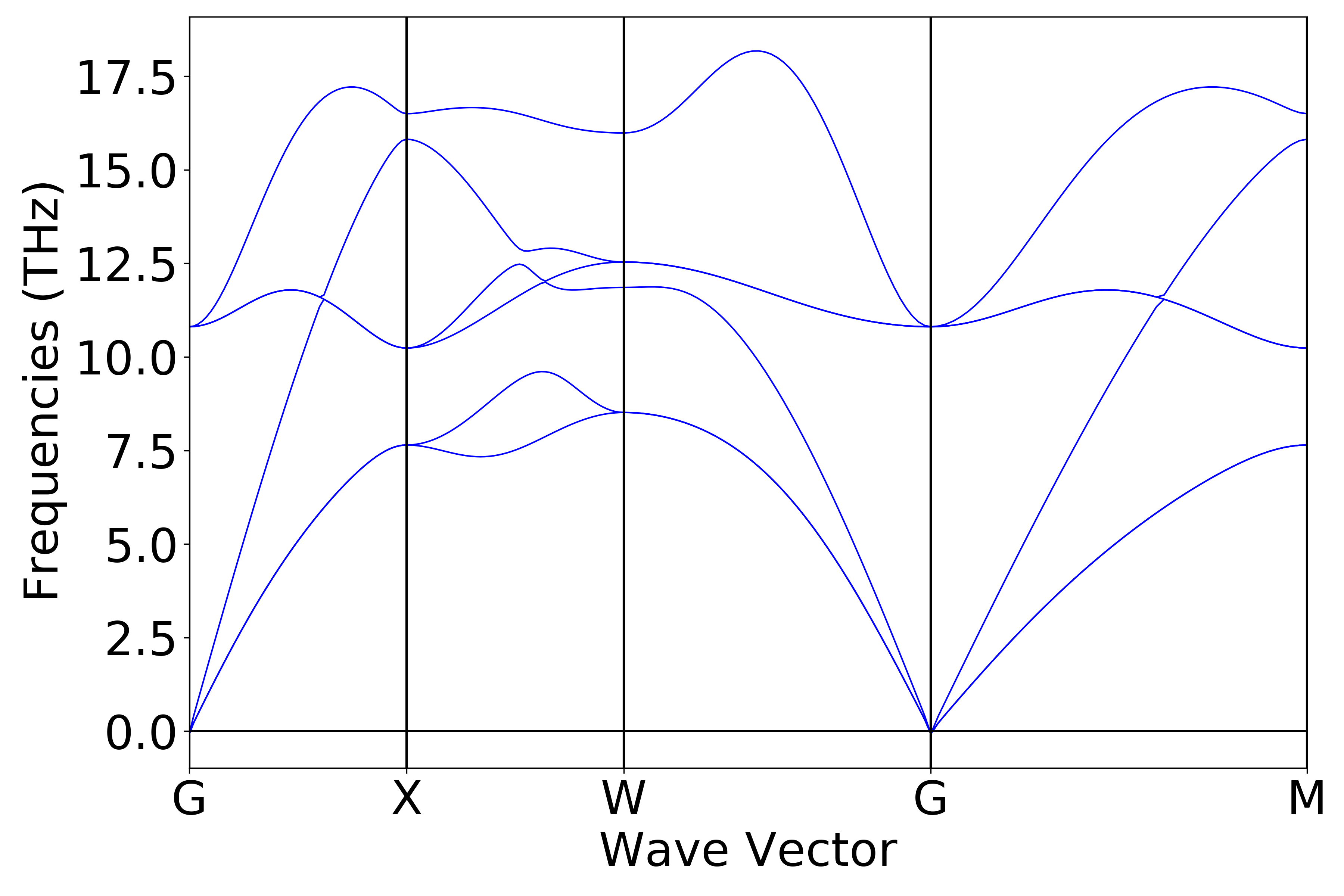
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
声子态密度数据处理
参考
9phonon_dosplot.py:执行代码可以得到类似以下声子态密度曲线:
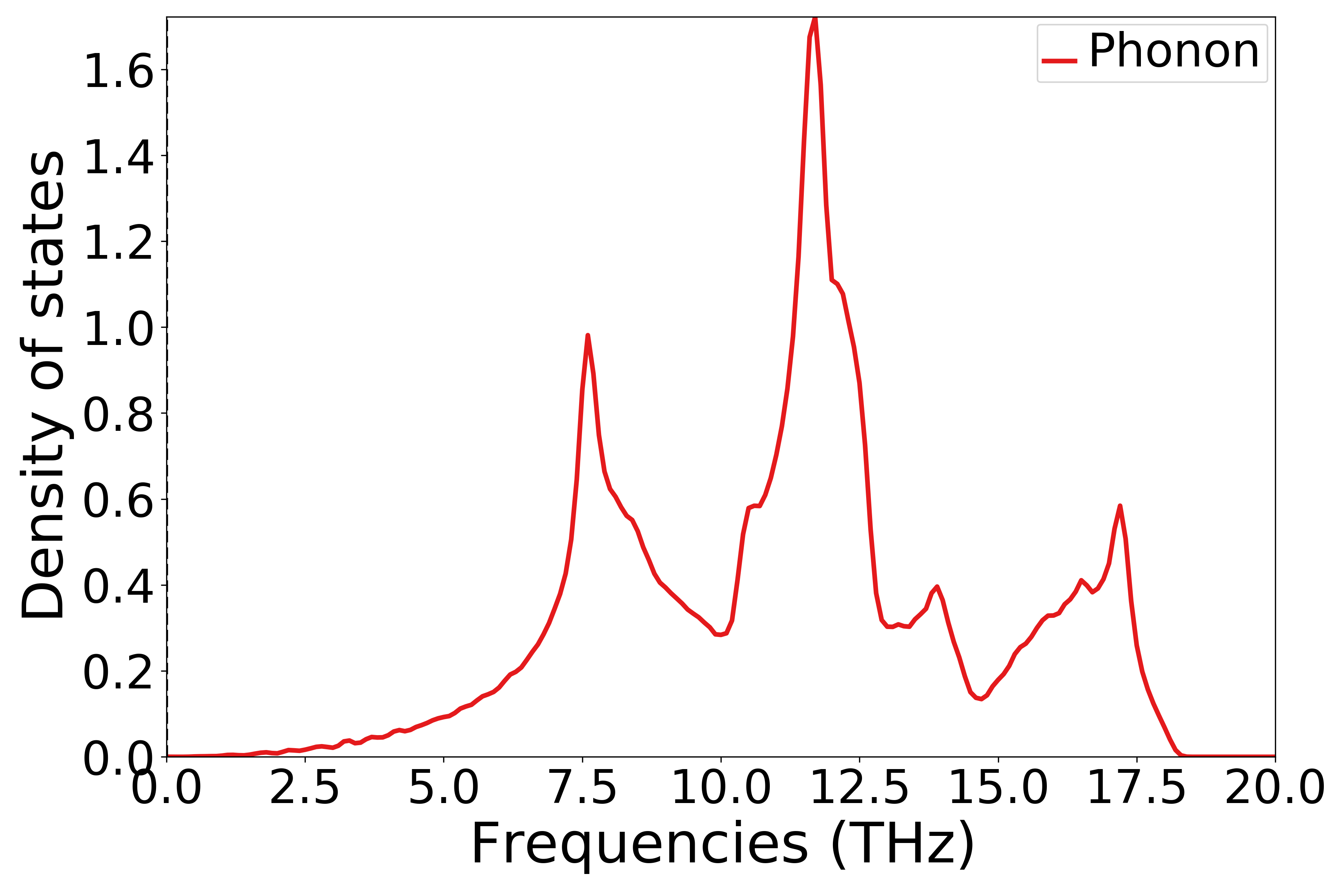
单晶硅声子态密度示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
声子热力学数据处理
可以参考 9phonon_thermal.py :
执行代码可以得到类似以下声子热力学曲线:
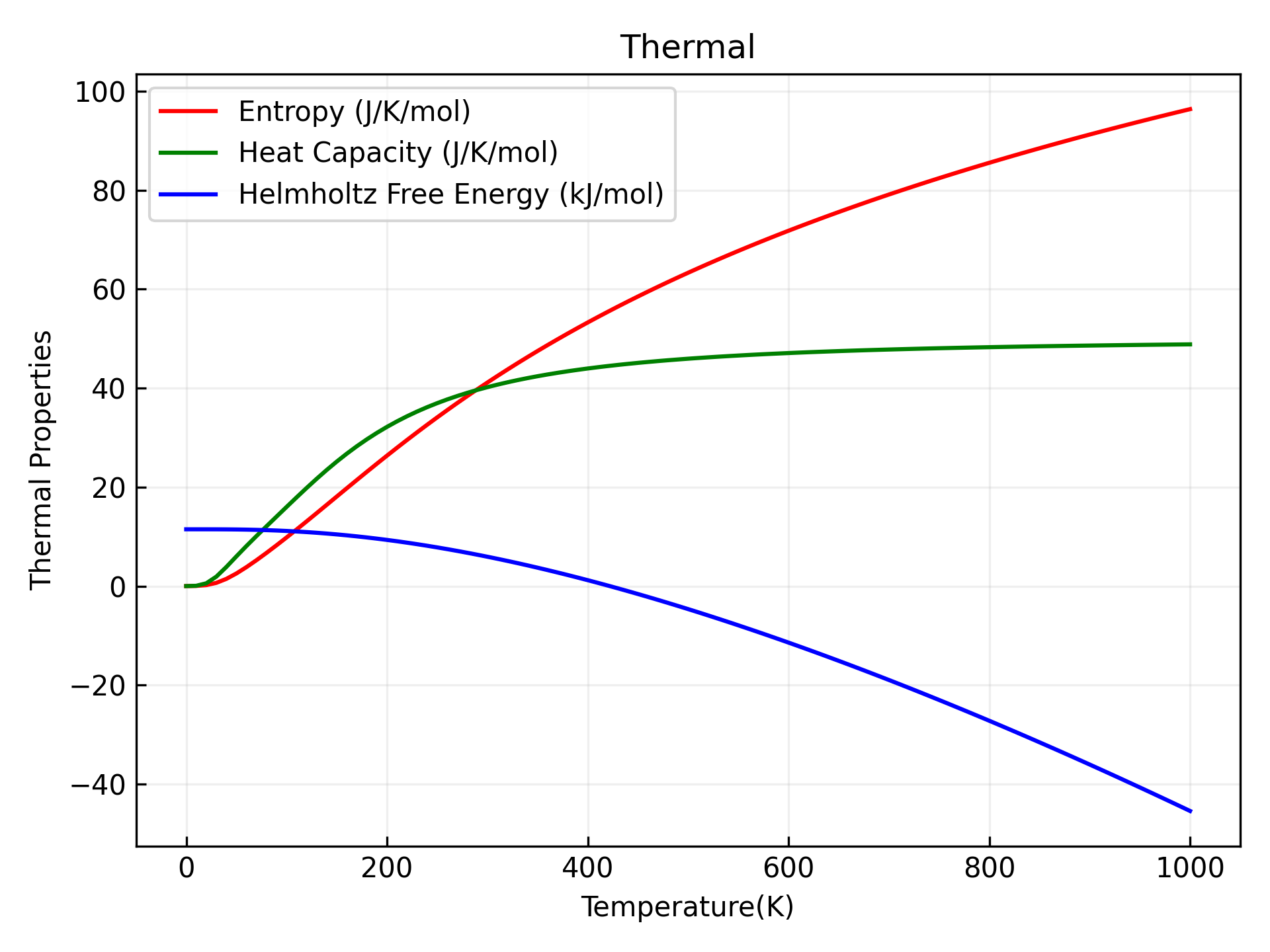
单晶硅声子热力学性质示意图
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.10 aimd分子动力学模拟数据处理
以快速入门 \(H_{2}O\) 分子体系分子动力学模拟得到的 aimd.h5 为例:
轨迹文件转换格式为.xyz或.dump
从aimd输出的hdf5文件中读取数据,并生成轨迹文件
生成的.xyz或.dump格式文件,可拖入OVITO观察,通过 Device Studio –> Simulator –> OVITO 打开 OVITO 可视化界面,将xyz文件或dump文件拖入OVITO即可。
参考 10write_aimd_traj.py :
执行代码将生成.xyz和.dump格式的轨迹文件,该文件可通过OVITO打开。关于结构文件转化的更多细节,请参考 structure结构转化
知识点:
OVITO 与 dspawpy 都不支持将非正交晶胞的体系保存为dump文件
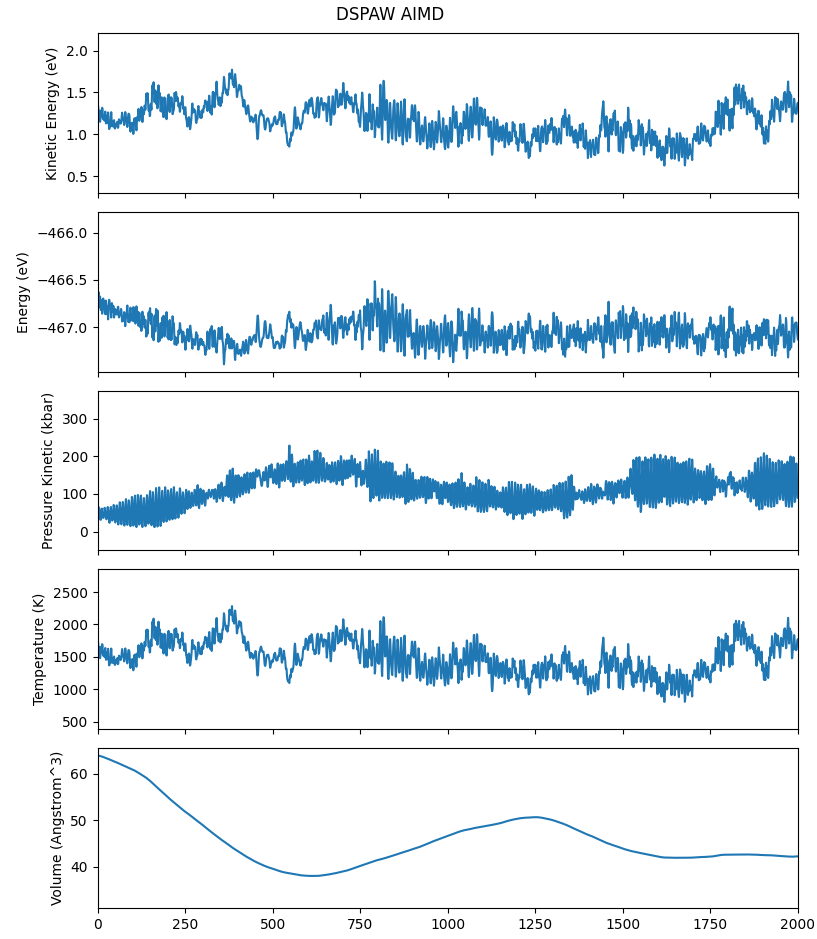
动力学过程中能量、温度等变化曲线
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
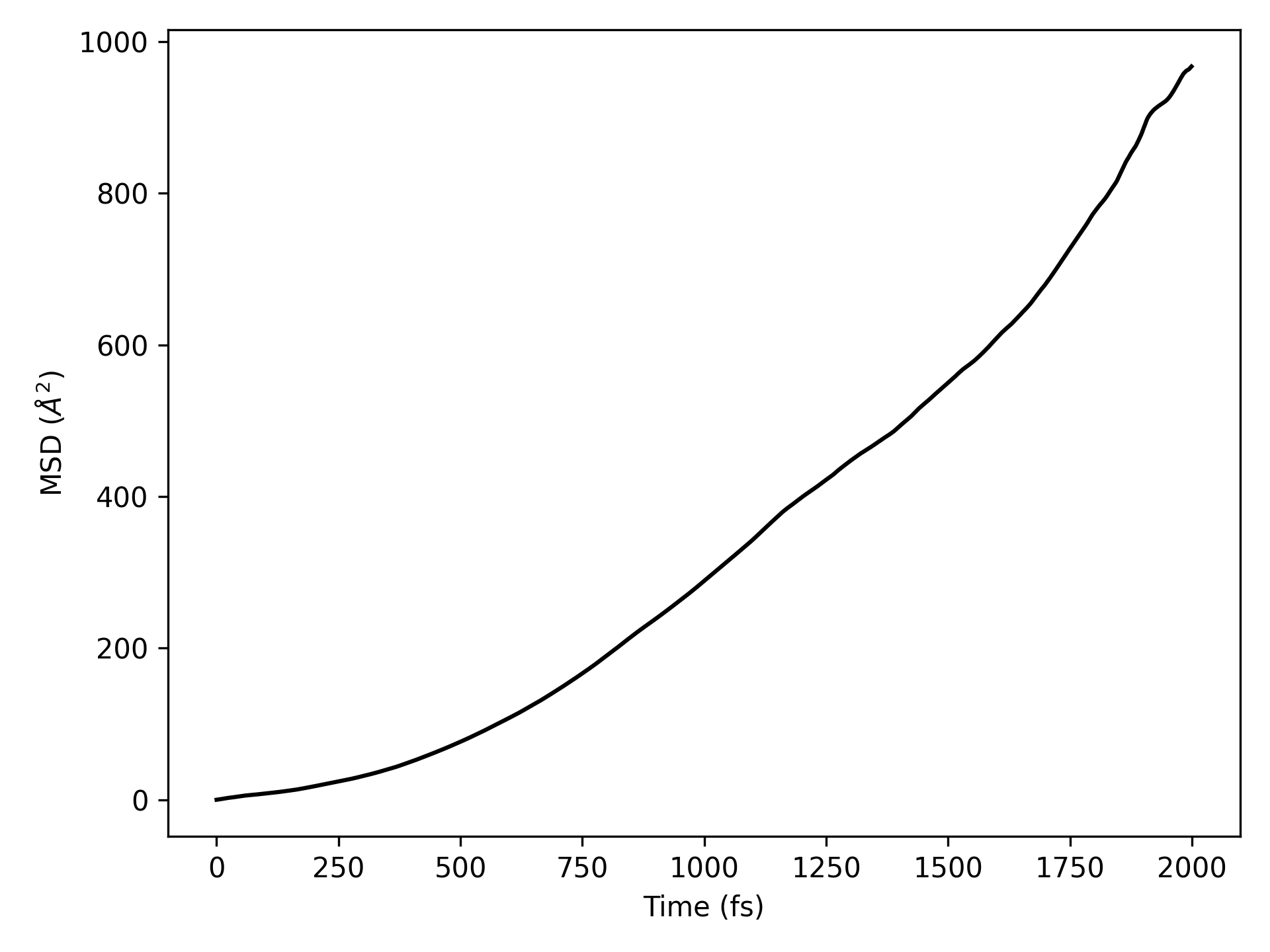
均方位移(MSD) 分析
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
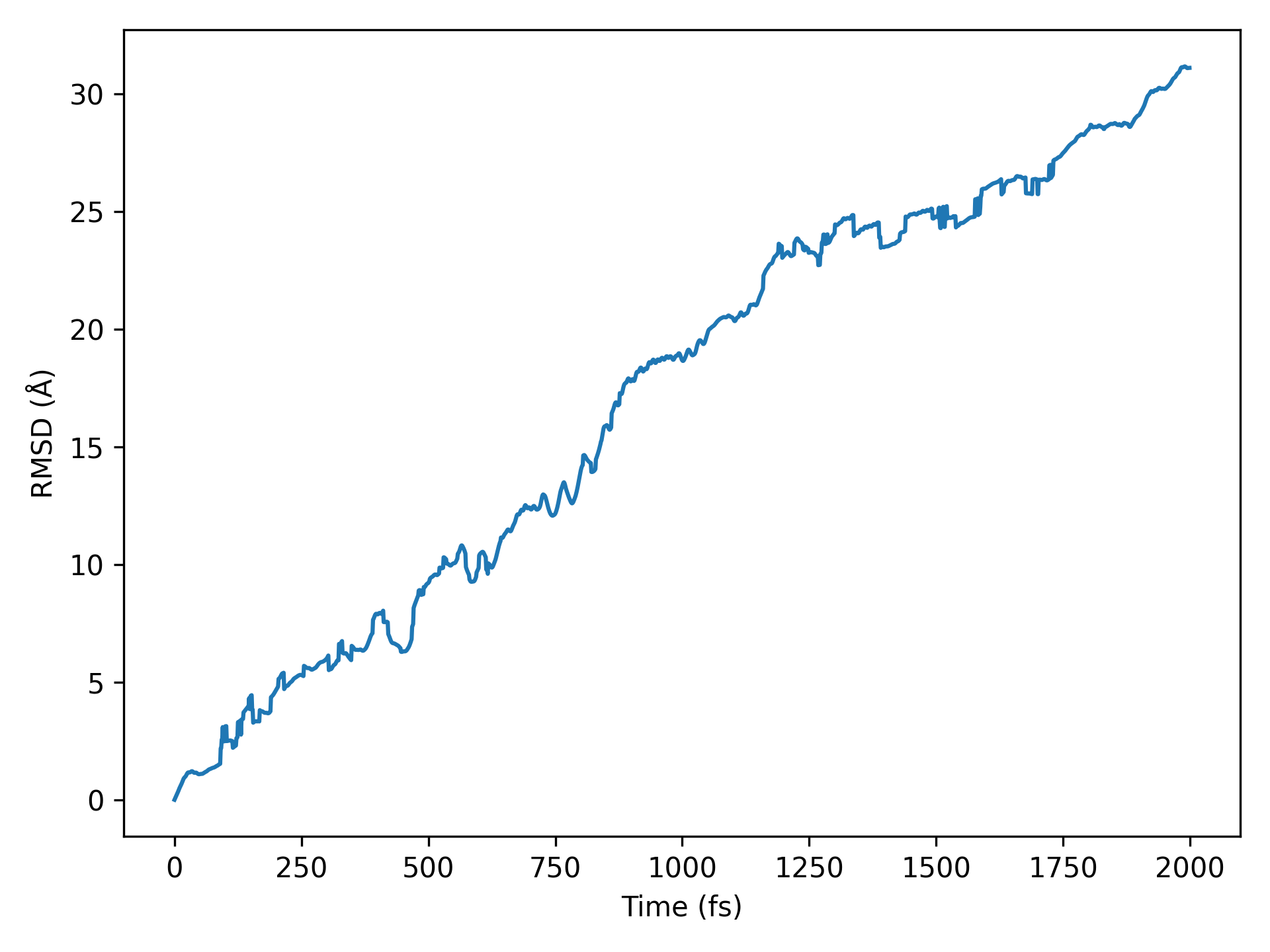
均方根偏差(RMSD) 分析
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
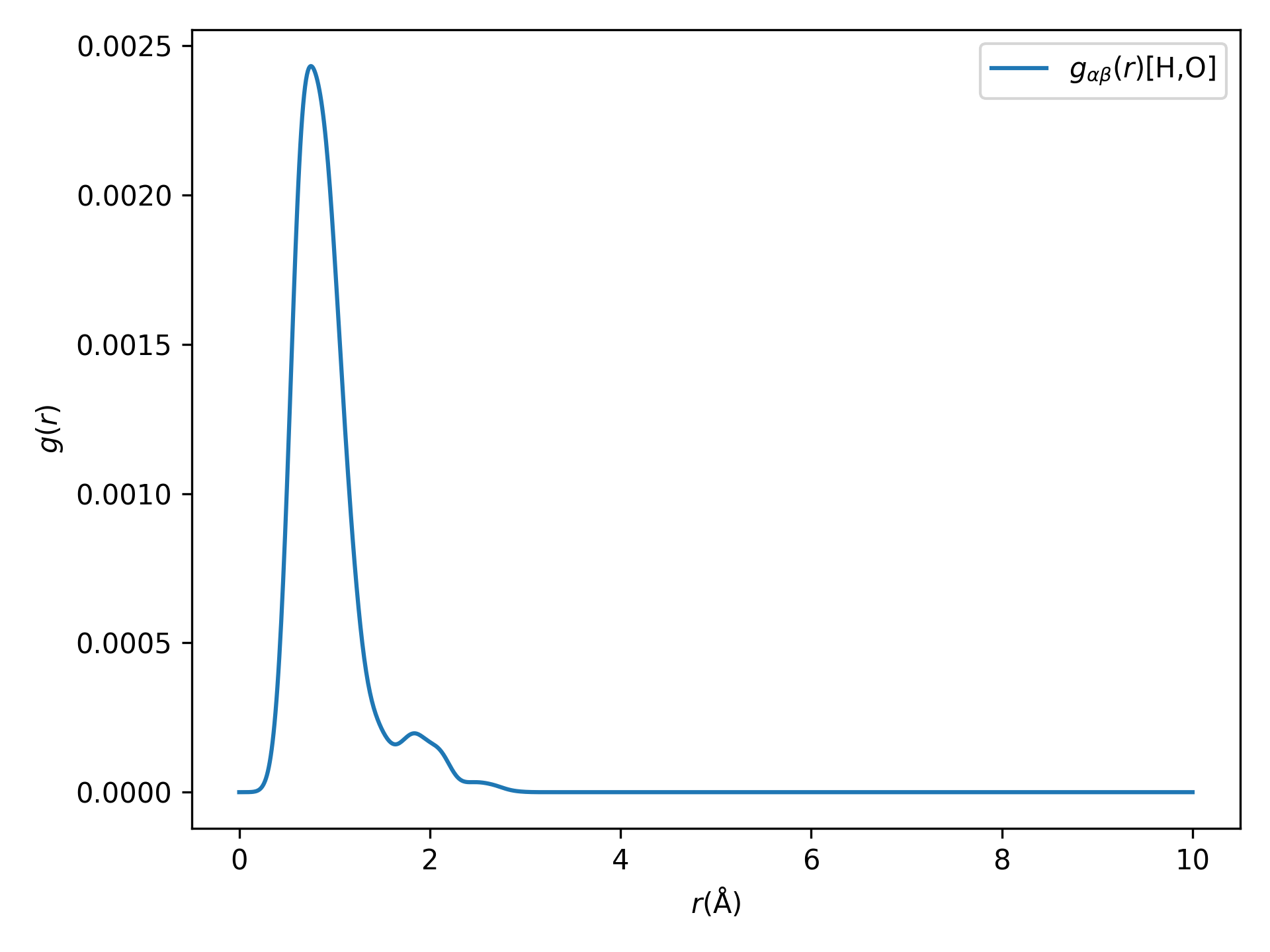
原子对分布函数或径向分布函数(RDFs) 分析
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.11 Polarization铁电极化数据处理
以快速入门 \(HfO_{2}\) 体系铁电计算得到的系列 scf.h5 为例:
参考
11Ferri.py:执行代码将生成如下组合图:
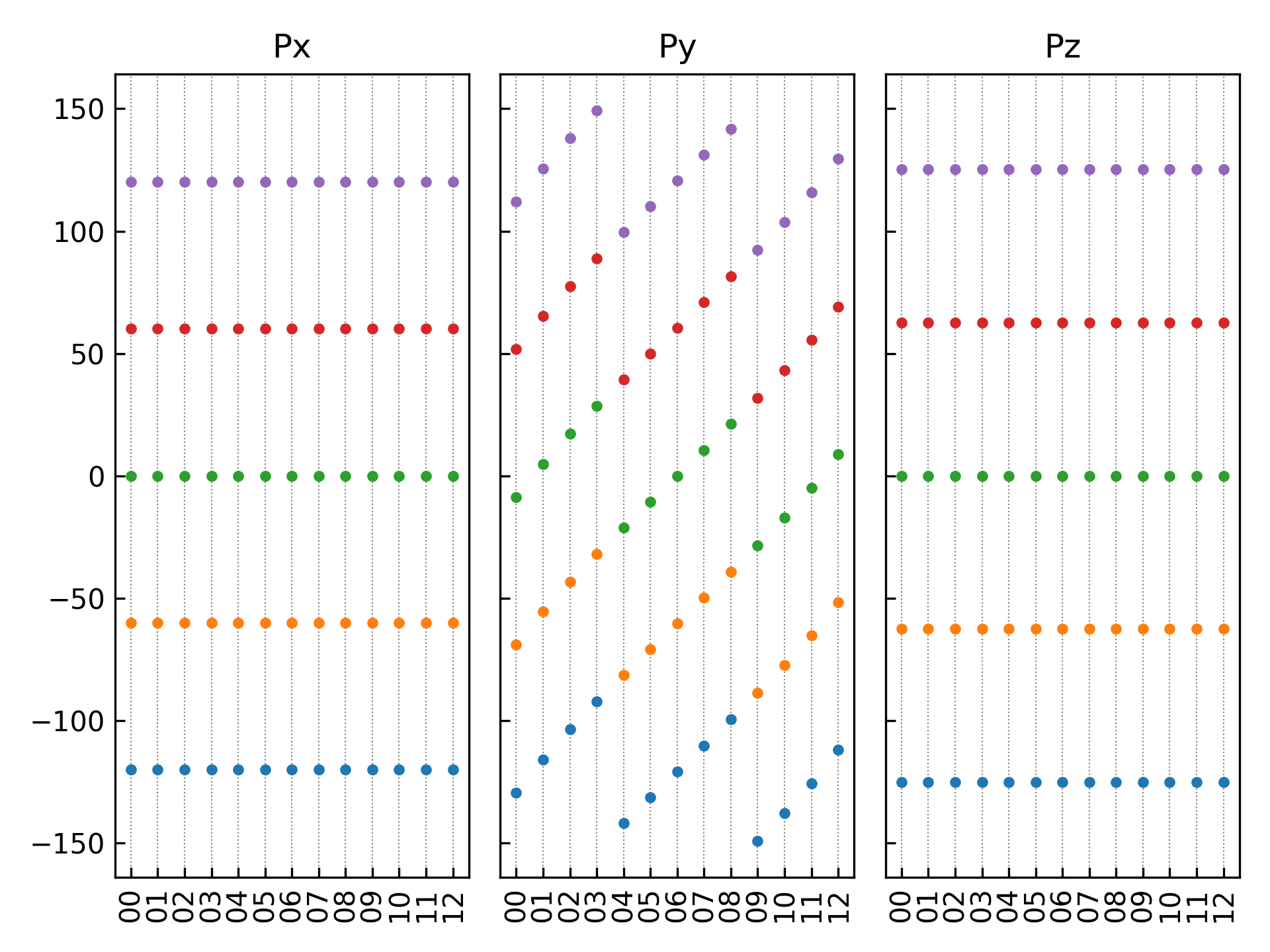
12组结构对应极化数值
查看首尾构型的铁电极化数值,可以参考如下:
1from dspawpy.plot import plot_polarization_figure 2 3plot_polarization_figure(directory='.', annotation=True, annotation_style=1) # 显示首尾构型的铁电极化数值
执行代码将生成如下组合图:
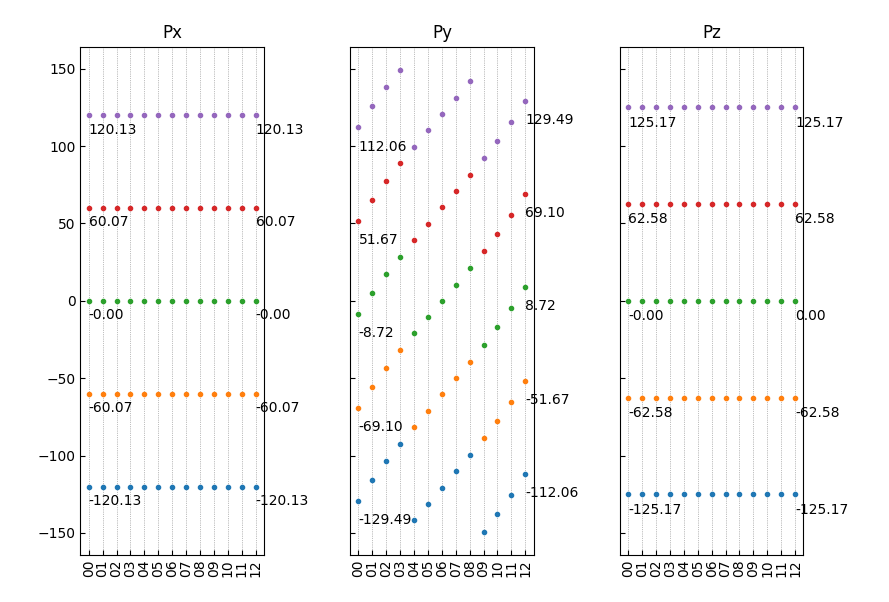
12组结构对应极化数值(带首尾构型数值)
也可以使用第二种批注样式:
1from dspawpy.plot import plot_polarization_figure 2 3plot_polarization_figure(directory='.', annotation=True, annotation_style=2) # 显示首尾构型的铁电极化数值
执行代码将生成如下组合图:
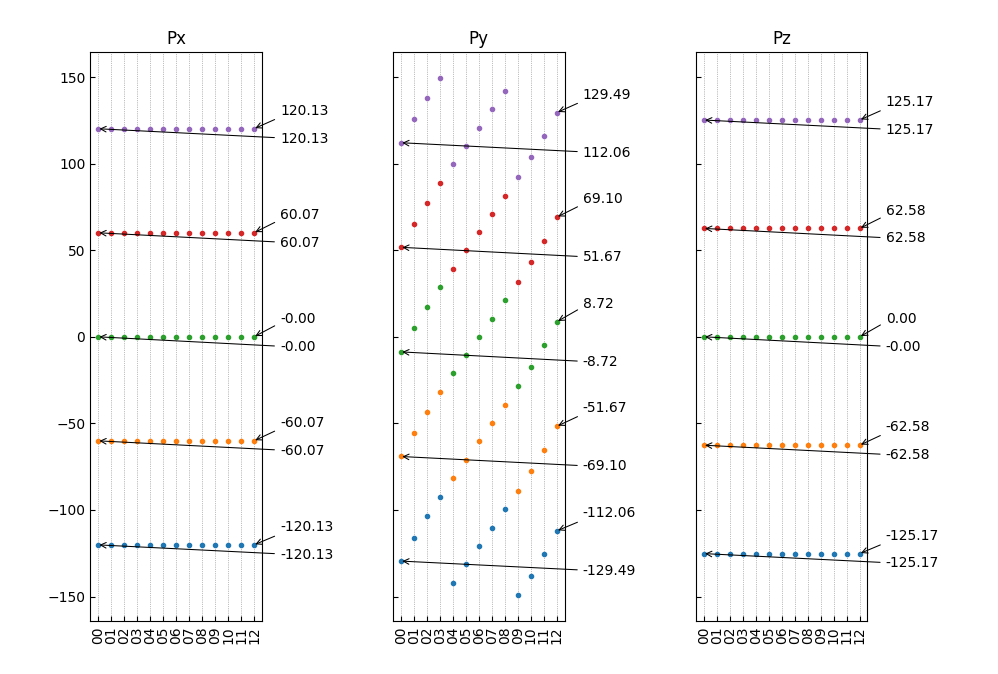
12组结构对应极化数值(带首尾构型数值)
警告
如果通过SSH连接到远程服务器执行上述脚本,出现QT相关的报错信息,可能是使用的程序(比如MobaXterm等)和QT库不兼容,要么更换程序(例如VSCode或者系统自带的终端命令行),要么请在python脚本第二行开始添加以下代码:
import matplotlib
matplotlib.use('agg')
8.12 ZPE零点振动能数据处理
以快速入门 \(CO\) 体系频率计算得到的 frequency.txt 文件为例,计算零点振动能,基于以下公式:
其中, \(\nu_i\) 是简正频率, \(h\) 是普朗克常数( \(6.626\times10^{-34} J\cdot s\)), \(N\) 是原子数。
参考
12getZPE.py:执行代码结果文件将保存到 ZPE.dat 文件中,文件内容如下:
Data read from D:\quickstart\2.13\frequency.txt Frequency (meV) 284.840038 --> Zero-point energy, ZPE (eV): 0.142420019
8.13 TS的热校正能
吸附质的熵变对能量的贡献
计算基于以下公式:
其中, \(\Delta S_{a d s}\) 表示吸附质的熵变,根据简谐近似计算。\(S_{v i b}\) 表示振动熵。 \(\nu_i\) 是简正频率,\(N_A\) 是阿伏伽德罗常数( \(6.022\times10^{23} mol^{-1}\) ), \(h\) 是普朗克常数( \(6.626\times10^{-34} J\cdot s\)), \(k_B\) 是玻尔兹曼常数( \(1.38\times10^{-23} J\cdot K^{-1}\)), \(R\) 是理想气体常数( \(8.314 J\cdot mol^{-1}\cdot K^{-1}\)), \(T\) 是体系温度,单位 \(K\)。
参考
13getTSads.py:执行代码结果文件将保存到 TS.dat 文件中,文件内容如下:
Data read from D:\quickstart\2.13\frequency.txt Frequency (THz) 68.873994 --> Entropy contribution, T*S (eV): 4.7566990201851275e-06
理想气体的熵变对能量的贡献
计算基于如下公式:
其中:
其中:\(I_A\) 到 \(I_C\) 是非线性分子的三个主惯性矩, \(I\) 是线性分子的简并惯性矩, \(\sigma\) 是分子的对称数。另外,monatomic 表示单原子分子,linear 表示线性分子,nonlinear 表示非线性分子。total spin 是总自旋数。 vib DOF 表示振动自由度。
参考
13getTSgas.py脚本处理:
8.14 附录
快速下载所有脚本,点击
用户脚本.zip